Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
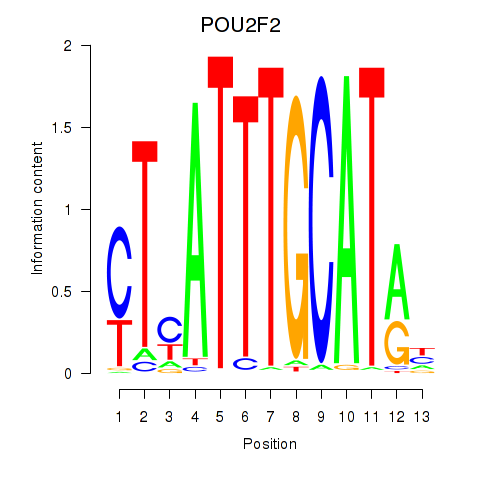
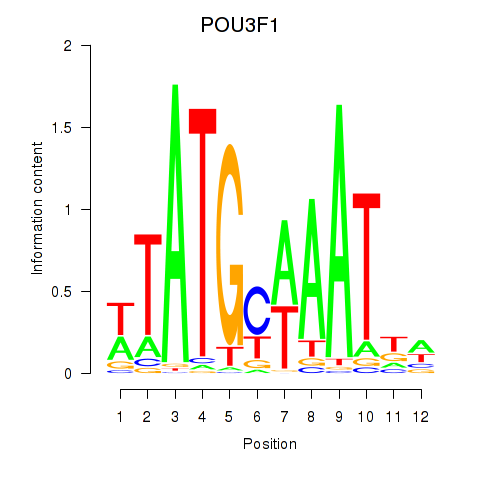
Results for POU2F2_POU3F1
Z-value: 1.21
Transcription factors associated with POU2F2_POU3F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU2F2
|
ENSG00000028277.16 | POU class 2 homeobox 2 |
|
POU3F1
|
ENSG00000185668.5 | POU class 3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU2F2 | hg19_v2_chr19_-_42636543_42636607 | 0.42 | 4.1e-01 | Click! |
| POU3F1 | hg19_v2_chr1_-_38512450_38512474 | -0.06 | 9.1e-01 | Click! |
Activity profile of POU2F2_POU3F1 motif
Sorted Z-values of POU2F2_POU3F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_26251835 | 2.00 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr17_+_38171681 | 1.90 |
ENST00000225474.2
ENST00000331769.2 ENST00000394148.3 ENST00000577675.1 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr17_+_38171614 | 1.71 |
ENST00000583218.1
ENST00000394149.3 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr2_+_89952792 | 1.20 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr1_+_149822620 | 1.17 |
ENST00000369159.2
|
HIST2H2AA4
|
histone cluster 2, H2aa4 |
| chr9_+_136325089 | 1.01 |
ENST00000291722.7
ENST00000316948.4 ENST00000540581.1 |
CACFD1
|
calcium channel flower domain containing 1 |
| chr1_-_153044083 | 0.99 |
ENST00000341611.2
|
SPRR2B
|
small proline-rich protein 2B |
| chr6_-_27860956 | 0.97 |
ENST00000359611.2
|
HIST1H2AM
|
histone cluster 1, H2am |
| chr1_-_149814478 | 0.92 |
ENST00000369161.3
|
HIST2H2AA3
|
histone cluster 2, H2aa3 |
| chr6_+_27861190 | 0.85 |
ENST00000303806.4
|
HIST1H2BO
|
histone cluster 1, H2bo |
| chr6_+_31588478 | 0.83 |
ENST00000376007.4
ENST00000376033.2 |
PRRC2A
|
proline-rich coiled-coil 2A |
| chr18_-_61311485 | 0.83 |
ENST00000436264.1
ENST00000356424.6 ENST00000341074.5 |
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 4 |
| chr19_-_17516449 | 0.80 |
ENST00000252593.6
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr9_-_35658007 | 0.76 |
ENST00000602361.1
|
RMRP
|
RNA component of mitochondrial RNA processing endoribonuclease |
| chr7_-_100183742 | 0.72 |
ENST00000310300.6
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr12_+_7053172 | 0.72 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr6_-_27782548 | 0.68 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr2_+_90248739 | 0.63 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr6_-_26124138 | 0.61 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr12_+_7052974 | 0.60 |
ENST00000544681.1
ENST00000537087.1 |
C12orf57
|
chromosome 12 open reading frame 57 |
| chr7_-_27153454 | 0.59 |
ENST00000522456.1
|
HOXA3
|
homeobox A3 |
| chr7_-_17500294 | 0.58 |
ENST00000439046.1
|
AC019117.2
|
AC019117.2 |
| chr12_+_122064398 | 0.58 |
ENST00000330079.7
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr6_+_27775899 | 0.57 |
ENST00000358739.3
|
HIST1H2AI
|
histone cluster 1, H2ai |
| chr5_+_159895275 | 0.55 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr6_-_27100529 | 0.54 |
ENST00000607124.1
ENST00000339812.2 ENST00000541790.1 |
HIST1H2BJ
|
histone cluster 1, H2bj |
| chr12_-_11175219 | 0.51 |
ENST00000390673.2
|
TAS2R19
|
taste receptor, type 2, member 19 |
| chr17_-_19651598 | 0.51 |
ENST00000570414.1
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr11_-_18258342 | 0.49 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr16_-_74808710 | 0.49 |
ENST00000219368.3
ENST00000544337.1 |
FA2H
|
fatty acid 2-hydroxylase |
| chr20_+_33292507 | 0.48 |
ENST00000414082.1
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr12_+_7053228 | 0.48 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chrX_-_24665353 | 0.47 |
ENST00000379144.2
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr1_+_32538520 | 0.47 |
ENST00000438825.1
ENST00000456834.2 ENST00000373634.4 ENST00000427288.1 |
TMEM39B
|
transmembrane protein 39B |
| chr19_+_56111680 | 0.47 |
ENST00000301073.3
|
ZNF524
|
zinc finger protein 524 |
| chr14_-_89960395 | 0.46 |
ENST00000555034.1
ENST00000553904.1 |
FOXN3
|
forkhead box N3 |
| chr17_+_62461569 | 0.46 |
ENST00000603557.1
ENST00000605096.1 |
MILR1
|
mast cell immunoglobulin-like receptor 1 |
| chr4_-_108204904 | 0.45 |
ENST00000510463.1
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr17_-_47841485 | 0.44 |
ENST00000506156.1
ENST00000240364.2 |
FAM117A
|
family with sequence similarity 117, member A |
| chr6_-_27114577 | 0.42 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chr19_-_1132207 | 0.42 |
ENST00000438103.2
|
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr12_-_10978957 | 0.42 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr1_+_32538492 | 0.42 |
ENST00000336294.5
|
TMEM39B
|
transmembrane protein 39B |
| chr7_-_41742697 | 0.41 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr6_-_10415218 | 0.41 |
ENST00000466073.1
ENST00000498450.1 |
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr6_+_26183958 | 0.41 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr12_+_113344582 | 0.41 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr14_+_65007177 | 0.40 |
ENST00000247207.6
|
HSPA2
|
heat shock 70kDa protein 2 |
| chr12_-_122238464 | 0.40 |
ENST00000546227.1
|
RHOF
|
ras homolog family member F (in filopodia) |
| chr12_+_122064673 | 0.39 |
ENST00000537188.1
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr8_+_102064237 | 0.38 |
ENST00000514926.1
|
RP11-302J23.1
|
RP11-302J23.1 |
| chr17_+_19091325 | 0.37 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr12_+_132312931 | 0.37 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr12_+_14927270 | 0.36 |
ENST00000544848.1
|
H2AFJ
|
H2A histone family, member J |
| chr11_+_110225855 | 0.36 |
ENST00000526605.1
ENST00000526703.1 |
RP11-347E10.1
|
RP11-347E10.1 |
| chr19_+_19144384 | 0.35 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr19_+_3572925 | 0.35 |
ENST00000333651.6
ENST00000417382.1 ENST00000453933.1 ENST00000262949.7 |
HMG20B
|
high mobility group 20B |
| chr12_+_113344811 | 0.34 |
ENST00000551241.1
ENST00000553185.1 ENST00000550689.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr19_+_41305085 | 0.34 |
ENST00000303961.4
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr2_+_90192768 | 0.34 |
ENST00000390275.2
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr19_+_11466062 | 0.33 |
ENST00000251473.5
ENST00000591329.1 ENST00000586380.1 |
DKFZP761J1410
|
Lipid phosphate phosphatase-related protein type 2 |
| chr5_-_141030943 | 0.32 |
ENST00000522783.1
ENST00000519800.1 ENST00000435817.2 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr5_-_88120083 | 0.32 |
ENST00000509373.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr19_+_41305330 | 0.31 |
ENST00000593972.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr6_-_10415470 | 0.31 |
ENST00000379604.2
ENST00000379613.3 |
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr10_+_24528108 | 0.30 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr19_-_44123734 | 0.30 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr12_-_66275350 | 0.30 |
ENST00000536648.1
|
RP11-366L20.2
|
Uncharacterized protein |
| chr8_-_33370607 | 0.29 |
ENST00000360742.5
ENST00000523305.1 |
TTI2
|
TELO2 interacting protein 2 |
| chr6_+_26217159 | 0.29 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chrX_-_55020511 | 0.29 |
ENST00000375006.3
ENST00000374992.2 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr19_-_41903161 | 0.29 |
ENST00000602129.1
ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5
|
exosome component 5 |
| chr3_-_87039662 | 0.29 |
ENST00000494229.1
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr19_-_893200 | 0.28 |
ENST00000269814.4
ENST00000395808.3 ENST00000312090.6 ENST00000325464.1 |
MED16
|
mediator complex subunit 16 |
| chr19_-_44124019 | 0.28 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr19_+_17858509 | 0.28 |
ENST00000594202.1
ENST00000252771.7 ENST00000389133.4 |
FCHO1
|
FCH domain only 1 |
| chr17_+_63096903 | 0.28 |
ENST00000582940.1
|
RP11-160O5.1
|
RP11-160O5.1 |
| chr11_-_62358972 | 0.27 |
ENST00000278279.3
|
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr17_+_40118805 | 0.26 |
ENST00000591072.1
ENST00000587679.1 ENST00000393888.1 ENST00000441615.2 |
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr17_+_40118773 | 0.26 |
ENST00000472031.1
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr19_-_19144243 | 0.26 |
ENST00000594445.1
ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2
|
SURP and G patch domain containing 2 |
| chr19_+_39903185 | 0.26 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr12_-_94673956 | 0.26 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr1_-_2458026 | 0.26 |
ENST00000435556.3
ENST00000378466.3 |
PANK4
|
pantothenate kinase 4 |
| chr19_-_50143452 | 0.25 |
ENST00000246792.3
|
RRAS
|
related RAS viral (r-ras) oncogene homolog |
| chr19_-_1021113 | 0.25 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr17_-_7832753 | 0.25 |
ENST00000303790.2
|
KCNAB3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
| chr19_-_40791302 | 0.25 |
ENST00000392038.2
ENST00000578123.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr1_-_27709793 | 0.25 |
ENST00000374027.3
ENST00000374025.3 |
CD164L2
|
CD164 sialomucin-like 2 |
| chr1_-_149783914 | 0.24 |
ENST00000369167.1
ENST00000427880.2 ENST00000545683.1 |
HIST2H2BF
|
histone cluster 2, H2bf |
| chr1_-_209792111 | 0.24 |
ENST00000455193.1
|
LAMB3
|
laminin, beta 3 |
| chr19_-_44171817 | 0.24 |
ENST00000593714.1
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr15_-_41166414 | 0.24 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr20_+_33292068 | 0.23 |
ENST00000374810.3
ENST00000374809.2 ENST00000451665.1 |
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr12_-_7245125 | 0.23 |
ENST00000542285.1
ENST00000540610.1 |
C1R
|
complement component 1, r subcomponent |
| chr19_-_47164386 | 0.23 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr19_+_19144666 | 0.23 |
ENST00000535288.1
ENST00000538663.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr19_-_50168962 | 0.23 |
ENST00000599223.1
ENST00000593922.1 ENST00000600022.1 ENST00000596765.1 ENST00000599144.1 ENST00000596822.1 ENST00000598108.1 ENST00000601373.1 ENST00000595034.1 ENST00000601291.1 |
IRF3
|
interferon regulatory factor 3 |
| chr11_+_64085560 | 0.22 |
ENST00000265462.4
ENST00000352435.4 ENST00000347941.4 |
PRDX5
|
peroxiredoxin 5 |
| chr1_+_32666188 | 0.22 |
ENST00000421922.2
|
CCDC28B
|
coiled-coil domain containing 28B |
| chr17_+_43213004 | 0.22 |
ENST00000586346.1
ENST00000398322.3 ENST00000592162.1 ENST00000376955.4 ENST00000321854.8 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr19_+_11466167 | 0.22 |
ENST00000591608.1
|
DKFZP761J1410
|
Lipid phosphate phosphatase-related protein type 2 |
| chr17_+_38497640 | 0.22 |
ENST00000394086.3
|
RARA
|
retinoic acid receptor, alpha |
| chr17_+_73521763 | 0.22 |
ENST00000167462.5
ENST00000375227.4 ENST00000392550.3 ENST00000578363.1 ENST00000579392.1 |
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr6_+_43139037 | 0.21 |
ENST00000265354.4
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor) |
| chr8_-_57123815 | 0.21 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr16_+_4666475 | 0.21 |
ENST00000591895.1
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr5_+_145317356 | 0.21 |
ENST00000511217.1
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr1_+_152975488 | 0.21 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr19_-_3971050 | 0.20 |
ENST00000545797.2
ENST00000596311.1 |
DAPK3
|
death-associated protein kinase 3 |
| chr19_+_17516624 | 0.20 |
ENST00000596322.1
ENST00000600008.1 ENST00000601885.1 |
CTD-2521M24.9
|
CTD-2521M24.9 |
| chr6_-_128222103 | 0.20 |
ENST00000434358.1
ENST00000543064.1 ENST00000368248.2 |
THEMIS
|
thymocyte selection associated |
| chr12_+_43086018 | 0.20 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr16_-_75285762 | 0.19 |
ENST00000564028.1
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr3_-_71632894 | 0.19 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr6_-_11232891 | 0.19 |
ENST00000379433.5
ENST00000379446.5 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr1_-_117021430 | 0.18 |
ENST00000423907.1
ENST00000434879.1 ENST00000443219.1 |
RP4-655J12.4
|
RP4-655J12.4 |
| chr15_+_45406519 | 0.18 |
ENST00000323030.5
|
DUOXA2
|
dual oxidase maturation factor 2 |
| chr12_+_130646999 | 0.18 |
ENST00000539839.1
ENST00000229030.4 |
FZD10
|
frizzled family receptor 10 |
| chr22_+_22764088 | 0.18 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr2_+_179318295 | 0.18 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr12_-_56224546 | 0.18 |
ENST00000357606.3
ENST00000547445.1 |
DNAJC14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr15_+_65337708 | 0.18 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr4_-_187517928 | 0.18 |
ENST00000512772.1
|
FAT1
|
FAT atypical cadherin 1 |
| chr6_+_26273144 | 0.18 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr1_+_156052354 | 0.18 |
ENST00000368301.2
|
LMNA
|
lamin A/C |
| chr11_+_537494 | 0.17 |
ENST00000270115.7
|
LRRC56
|
leucine rich repeat containing 56 |
| chr11_-_62359027 | 0.17 |
ENST00000494385.1
ENST00000308436.7 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr19_+_16607122 | 0.17 |
ENST00000221671.3
ENST00000594035.1 ENST00000599550.1 ENST00000594813.1 |
C19orf44
|
chromosome 19 open reading frame 44 |
| chr5_+_158527630 | 0.17 |
ENST00000523301.1
|
RP11-175K6.1
|
RP11-175K6.1 |
| chr6_+_34204642 | 0.17 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr5_-_175965008 | 0.16 |
ENST00000537487.1
|
RNF44
|
ring finger protein 44 |
| chr11_-_568369 | 0.16 |
ENST00000534540.1
ENST00000528245.1 ENST00000500447.1 ENST00000533920.1 |
MIR210HG
|
MIR210 host gene (non-protein coding) |
| chr17_-_19651654 | 0.16 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr9_+_35658262 | 0.16 |
ENST00000378407.3
ENST00000378406.1 ENST00000426546.2 ENST00000327351.2 ENST00000421582.2 |
CCDC107
|
coiled-coil domain containing 107 |
| chr12_-_58212487 | 0.16 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr3_-_46068969 | 0.16 |
ENST00000542109.1
ENST00000395946.2 |
XCR1
|
chemokine (C motif) receptor 1 |
| chr5_-_88120151 | 0.15 |
ENST00000506716.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr11_-_78140882 | 0.15 |
ENST00000513207.2
|
RP11-452H21.4
|
RP11-452H21.4 |
| chr16_-_75285380 | 0.15 |
ENST00000393420.6
ENST00000162330.5 |
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr19_-_45982076 | 0.15 |
ENST00000423698.2
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr2_+_47922958 | 0.15 |
ENST00000606499.1
|
MSH6
|
mutS homolog 6 |
| chr2_-_61108449 | 0.15 |
ENST00000439412.1
ENST00000452343.1 |
AC010733.4
|
AC010733.4 |
| chr7_+_16793160 | 0.14 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr19_+_42788172 | 0.14 |
ENST00000160740.3
|
CIC
|
capicua transcriptional repressor |
| chr12_+_93096619 | 0.14 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr14_+_93389425 | 0.14 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chr16_+_56691606 | 0.14 |
ENST00000334350.6
|
MT1F
|
metallothionein 1F |
| chr15_-_45406348 | 0.14 |
ENST00000603300.1
|
DUOX2
|
dual oxidase 2 |
| chr2_-_74555350 | 0.14 |
ENST00000444570.1
|
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr17_-_73775839 | 0.14 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr4_+_40194609 | 0.13 |
ENST00000508513.1
|
RHOH
|
ras homolog family member H |
| chr4_-_40516560 | 0.13 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr10_-_29811456 | 0.13 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr3_-_126327398 | 0.13 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr2_+_68962014 | 0.13 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr6_+_26158343 | 0.13 |
ENST00000377777.4
ENST00000289316.2 |
HIST1H2BD
|
histone cluster 1, H2bd |
| chr9_+_136325149 | 0.13 |
ENST00000542192.1
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr4_-_110223799 | 0.13 |
ENST00000399132.1
ENST00000399126.1 ENST00000505591.1 |
COL25A1
|
collagen, type XXV, alpha 1 |
| chr19_+_1269324 | 0.13 |
ENST00000589710.1
ENST00000588230.1 ENST00000413636.2 ENST00000586472.1 ENST00000589686.1 ENST00000444172.2 ENST00000587323.1 ENST00000320936.5 ENST00000587896.1 ENST00000589235.1 ENST00000591659.1 |
CIRBP
|
cold inducible RNA binding protein |
| chr12_-_51611477 | 0.13 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr21_+_17566643 | 0.12 |
ENST00000419952.1
ENST00000445461.2 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr19_-_19281054 | 0.12 |
ENST00000424583.2
ENST00000410050.1 ENST00000409224.1 ENST00000409447.2 |
MEF2B
|
myocyte enhancer factor 2B |
| chr15_+_67458357 | 0.12 |
ENST00000537194.2
|
SMAD3
|
SMAD family member 3 |
| chr16_-_30107491 | 0.12 |
ENST00000566134.1
ENST00000565110.1 ENST00000398841.1 ENST00000398838.4 |
YPEL3
|
yippee-like 3 (Drosophila) |
| chr17_-_36358166 | 0.12 |
ENST00000537432.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr12_-_62653903 | 0.12 |
ENST00000552075.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr6_-_53013620 | 0.12 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr17_+_80858418 | 0.12 |
ENST00000574422.1
|
TBCD
|
tubulin folding cofactor D |
| chr2_+_152214098 | 0.12 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr16_-_19897455 | 0.12 |
ENST00000568214.1
ENST00000569479.1 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr18_-_30716038 | 0.12 |
ENST00000581852.1
|
CCDC178
|
coiled-coil domain containing 178 |
| chr2_-_214013353 | 0.12 |
ENST00000451136.2
ENST00000421754.2 ENST00000374327.4 ENST00000413091.3 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr1_-_6052463 | 0.11 |
ENST00000378156.4
|
NPHP4
|
nephronophthisis 4 |
| chr10_-_103880209 | 0.11 |
ENST00000425280.1
|
LDB1
|
LIM domain binding 1 |
| chr12_+_93096759 | 0.11 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr3_+_50192537 | 0.11 |
ENST00000002829.3
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr10_-_98031155 | 0.11 |
ENST00000495266.1
|
BLNK
|
B-cell linker |
| chr19_-_40791160 | 0.11 |
ENST00000358335.5
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr17_+_40118759 | 0.11 |
ENST00000393892.3
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr15_-_45406385 | 0.11 |
ENST00000389039.6
|
DUOX2
|
dual oxidase 2 |
| chr2_-_225811747 | 0.11 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr4_+_124320665 | 0.11 |
ENST00000394339.2
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr9_+_35658377 | 0.10 |
ENST00000378409.3
|
CCDC107
|
coiled-coil domain containing 107 |
| chr5_-_2751762 | 0.10 |
ENST00000302057.5
ENST00000382611.6 |
IRX2
|
iroquois homeobox 2 |
| chr12_+_19358192 | 0.10 |
ENST00000538305.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chrX_-_154688276 | 0.10 |
ENST00000369445.2
|
F8A3
|
coagulation factor VIII-associated 3 |
| chr2_+_207804278 | 0.10 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr3_+_124303539 | 0.10 |
ENST00000428018.2
|
KALRN
|
kalirin, RhoGEF kinase |
| chr22_+_38093005 | 0.10 |
ENST00000406386.3
|
TRIOBP
|
TRIO and F-actin binding protein |
| chr12_-_7245080 | 0.10 |
ENST00000541042.1
ENST00000540242.1 |
C1R
|
complement component 1, r subcomponent |
| chr17_-_7307358 | 0.10 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr12_+_72080253 | 0.10 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr2_-_166930131 | 0.10 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr19_-_47291843 | 0.10 |
ENST00000542575.2
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr3_-_87040259 | 0.10 |
ENST00000383698.3
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr3_-_192445289 | 0.10 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr18_-_53089538 | 0.10 |
ENST00000566777.1
|
TCF4
|
transcription factor 4 |
| chr3_+_187957646 | 0.10 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr16_-_73082274 | 0.10 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU2F2_POU3F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.3 | 3.6 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.3 | 0.8 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.3 | 0.8 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) |
| 0.1 | 0.7 | GO:0021623 | optic cup structural organization(GO:0003409) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.1 | 0.4 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.4 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 1.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.6 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.2 | GO:0061145 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.1 | 0.5 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.3 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.4 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.1 | GO:1900738 | dense core granule biogenesis(GO:0061110) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of muscle(GO:1901079) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 1.0 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.5 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.5 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.3 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.8 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0042335 | cuticle development(GO:0042335) hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.4 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.0 | GO:1900200 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.3 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.2 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.2 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:1900005 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.2 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.4 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.2 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.4 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:0061054 | negative regulation of mesodermal cell fate specification(GO:0042662) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.0 | 0.0 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 1.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 3.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 1.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0035547 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.0 | GO:1905069 | allantois development(GO:1905069) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.2 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.2 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.2 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.2 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.3 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.2 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 4.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 0.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 1.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.4 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 1.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.3 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.7 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 1.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.6 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.2 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.1 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.1 | GO:0004877 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.2 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 3.4 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 1.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 4.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |