Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
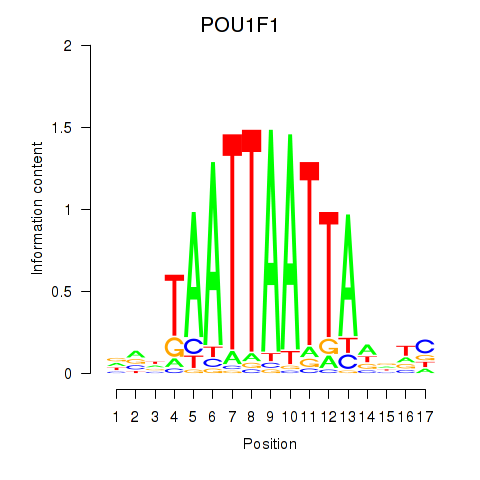
Results for POU1F1
Z-value: 0.44
Transcription factors associated with POU1F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU1F1
|
ENSG00000064835.6 | POU class 1 homeobox 1 |
Activity profile of POU1F1 motif
Sorted Z-values of POU1F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_78359999 | 0.37 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr7_-_35013217 | 0.36 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr9_-_95055923 | 0.34 |
ENST00000430417.1
|
IARS
|
isoleucyl-tRNA synthetase |
| chr7_+_13141010 | 0.33 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr17_-_57158523 | 0.30 |
ENST00000581468.1
|
TRIM37
|
tripartite motif containing 37 |
| chr3_-_195076933 | 0.29 |
ENST00000423531.1
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr5_+_147691979 | 0.29 |
ENST00000274565.4
|
SPINK7
|
serine peptidase inhibitor, Kazal type 7 (putative) |
| chr5_+_136070614 | 0.26 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr21_-_43735628 | 0.25 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr6_-_52859046 | 0.24 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr3_+_38537960 | 0.23 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr14_+_39944025 | 0.22 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chrX_+_150565038 | 0.22 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr6_-_134861089 | 0.22 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr2_-_165630264 | 0.20 |
ENST00000452626.1
|
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr4_-_112993808 | 0.20 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr5_+_115177178 | 0.20 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr12_+_38710555 | 0.19 |
ENST00000551464.1
|
ALG10B
|
ALG10B, alpha-1,2-glucosyltransferase |
| chr3_+_138340067 | 0.19 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr14_-_38036271 | 0.19 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr2_-_17981462 | 0.19 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr2_+_67624430 | 0.18 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr12_+_64798095 | 0.15 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr15_-_56757329 | 0.15 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr10_+_94352956 | 0.15 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr4_+_25162253 | 0.14 |
ENST00000512921.1
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta |
| chr12_-_10282742 | 0.13 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr20_+_5986727 | 0.13 |
ENST00000378863.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr10_-_115904361 | 0.13 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr4_-_146261573 | 0.13 |
ENST00000610239.1
|
RP11-142A22.4
|
RP11-142A22.4 |
| chr9_+_125133467 | 0.13 |
ENST00000426608.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr12_+_69753448 | 0.13 |
ENST00000247843.2
ENST00000548020.1 ENST00000549685.1 ENST00000552955.1 |
YEATS4
|
YEATS domain containing 4 |
| chr7_+_134528635 | 0.13 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chrX_+_150565653 | 0.12 |
ENST00000330374.6
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr16_-_28937027 | 0.12 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr14_+_55494323 | 0.12 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr7_+_107224364 | 0.12 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr4_-_118006697 | 0.11 |
ENST00000310754.4
|
TRAM1L1
|
translocation associated membrane protein 1-like 1 |
| chr2_-_207078086 | 0.11 |
ENST00000442134.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr15_+_58702742 | 0.11 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr6_+_114178512 | 0.11 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr15_-_75748143 | 0.11 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr20_+_5987890 | 0.11 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr4_-_153601136 | 0.11 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr1_+_192127578 | 0.11 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr15_+_101417919 | 0.11 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr6_-_52859968 | 0.11 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr14_+_31046959 | 0.11 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr17_+_78518617 | 0.11 |
ENST00000537330.1
ENST00000570891.1 |
RPTOR
|
regulatory associated protein of MTOR, complex 1 |
| chr8_+_92114060 | 0.10 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr9_-_21482312 | 0.10 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr5_+_59783941 | 0.10 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr3_-_108248169 | 0.10 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr3_-_96337000 | 0.10 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr11_-_63376013 | 0.10 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr20_+_5986756 | 0.10 |
ENST00000452938.1
|
CRLS1
|
cardiolipin synthase 1 |
| chr4_+_74301880 | 0.10 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr19_-_44388116 | 0.10 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chrX_+_84258832 | 0.09 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr12_-_67197760 | 0.09 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr12_-_71148413 | 0.09 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr7_+_99425633 | 0.09 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr3_+_108308845 | 0.09 |
ENST00000479138.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr14_+_61449076 | 0.09 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr16_-_2379688 | 0.08 |
ENST00000567910.1
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3 |
| chr16_+_15489629 | 0.08 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr4_+_189321881 | 0.08 |
ENST00000512839.1
ENST00000513313.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
| chr2_-_207078154 | 0.08 |
ENST00000447845.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr10_+_94594351 | 0.08 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr11_-_22647350 | 0.08 |
ENST00000327470.3
|
FANCF
|
Fanconi anemia, complementation group F |
| chr7_-_140482926 | 0.08 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr3_+_136649311 | 0.08 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr17_+_1944790 | 0.08 |
ENST00000575162.1
|
DPH1
|
diphthamide biosynthesis 1 |
| chr12_+_59194154 | 0.08 |
ENST00000548969.1
|
RP11-362K2.2
|
Protein LOC100506869 |
| chr9_+_12693336 | 0.08 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chrX_-_102531717 | 0.08 |
ENST00000372680.1
|
TCEAL5
|
transcription elongation factor A (SII)-like 5 |
| chr16_-_18923035 | 0.07 |
ENST00000563836.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr5_+_95066823 | 0.07 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr12_-_49581152 | 0.07 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr4_+_41937131 | 0.07 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr4_+_95128748 | 0.07 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr8_-_79470728 | 0.07 |
ENST00000522807.1
ENST00000519242.1 ENST00000522302.1 |
RP11-594N15.2
|
RP11-594N15.2 |
| chr7_-_112758665 | 0.07 |
ENST00000397764.3
|
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr14_+_101295638 | 0.07 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr3_-_20053741 | 0.07 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr1_+_76251912 | 0.07 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr14_+_55493920 | 0.07 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr2_-_152118352 | 0.07 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr16_-_66764119 | 0.07 |
ENST00000569320.1
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr9_-_5833027 | 0.07 |
ENST00000339450.5
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr16_+_21623392 | 0.07 |
ENST00000562961.1
|
METTL9
|
methyltransferase like 9 |
| chr14_-_92247032 | 0.07 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr7_-_112758589 | 0.07 |
ENST00000413744.1
ENST00000439551.1 ENST00000441359.1 |
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr8_-_57233103 | 0.07 |
ENST00000303749.3
ENST00000522671.1 |
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5 |
| chr19_+_19144384 | 0.07 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr2_-_190446738 | 0.07 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr10_-_14050522 | 0.06 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr2_+_172309634 | 0.06 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr2_-_109605663 | 0.06 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr5_+_179135246 | 0.06 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr6_+_55192267 | 0.06 |
ENST00000340465.2
|
GFRAL
|
GDNF family receptor alpha like |
| chr4_+_3344141 | 0.06 |
ENST00000306648.7
|
RGS12
|
regulator of G-protein signaling 12 |
| chr4_+_37455536 | 0.06 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr4_+_48833234 | 0.06 |
ENST00000510824.1
ENST00000425583.2 |
OCIAD1
|
OCIA domain containing 1 |
| chr2_+_103089756 | 0.06 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr17_-_6524159 | 0.06 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr9_+_125132803 | 0.06 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr10_-_28571015 | 0.06 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr6_+_29555683 | 0.06 |
ENST00000383640.2
|
OR2H2
|
olfactory receptor, family 2, subfamily H, member 2 |
| chr19_+_21579958 | 0.06 |
ENST00000339914.6
ENST00000599461.1 |
ZNF493
|
zinc finger protein 493 |
| chr10_-_75226166 | 0.06 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr15_-_72521017 | 0.06 |
ENST00000561609.1
|
PKM
|
pyruvate kinase, muscle |
| chr17_+_46189311 | 0.06 |
ENST00000582481.1
|
SNX11
|
sorting nexin 11 |
| chr18_-_13915530 | 0.06 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr3_+_145782358 | 0.06 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chr2_-_152118276 | 0.05 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr10_+_17270214 | 0.05 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr17_+_40950797 | 0.05 |
ENST00000588408.1
ENST00000585355.1 |
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr6_+_26087509 | 0.05 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr3_+_44626359 | 0.05 |
ENST00000412641.1
|
ZNF197
|
zinc finger protein 197 |
| chr10_+_57358750 | 0.05 |
ENST00000512524.2
|
MTRNR2L5
|
MT-RNR2-like 5 |
| chr3_+_82035289 | 0.05 |
ENST00000470263.1
ENST00000494340.1 |
RP11-260O18.1
|
RP11-260O18.1 |
| chr5_-_79946820 | 0.05 |
ENST00000604882.1
|
MTRNR2L2
|
MT-RNR2-like 2 |
| chr4_-_46996424 | 0.05 |
ENST00000264318.3
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr16_-_70835034 | 0.05 |
ENST00000261776.5
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr11_-_111649074 | 0.05 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr19_+_49199209 | 0.05 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr2_+_208414985 | 0.05 |
ENST00000414681.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr6_-_138833630 | 0.05 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr1_-_54411255 | 0.05 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr14_+_89060749 | 0.05 |
ENST00000555900.1
ENST00000406216.3 ENST00000557737.1 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr6_-_110501126 | 0.05 |
ENST00000368938.1
|
WASF1
|
WAS protein family, member 1 |
| chr11_-_26593677 | 0.05 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr6_+_26440700 | 0.05 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chrX_-_77225135 | 0.05 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr4_-_74486109 | 0.04 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr10_+_18549645 | 0.04 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr7_-_84122033 | 0.04 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr9_+_125133315 | 0.04 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr2_+_161993412 | 0.04 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr7_+_116654958 | 0.04 |
ENST00000449366.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr5_+_129083772 | 0.04 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr17_+_66255310 | 0.04 |
ENST00000448504.2
|
ARSG
|
arylsulfatase G |
| chr8_-_30670384 | 0.04 |
ENST00000221138.4
ENST00000518243.1 |
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr1_+_111682058 | 0.04 |
ENST00000545121.1
|
CEPT1
|
choline/ethanolamine phosphotransferase 1 |
| chr12_-_22063787 | 0.04 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr12_+_34175398 | 0.04 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr14_+_62164340 | 0.04 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr17_+_29664830 | 0.04 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chrX_-_71458802 | 0.04 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr3_+_138340049 | 0.04 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr18_-_59274139 | 0.04 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chr19_+_50016610 | 0.04 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chrX_-_73061339 | 0.04 |
ENST00000602863.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr12_+_133066137 | 0.04 |
ENST00000434748.2
|
FBRSL1
|
fibrosin-like 1 |
| chr8_-_141774467 | 0.04 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr18_+_61445205 | 0.04 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr1_+_234765057 | 0.04 |
ENST00000429269.1
|
LINC00184
|
long intergenic non-protein coding RNA 184 |
| chr12_+_26348429 | 0.04 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr17_+_7155819 | 0.04 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr11_-_4719072 | 0.04 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr1_+_93297622 | 0.04 |
ENST00000315741.5
|
RPL5
|
ribosomal protein L5 |
| chr9_-_95055956 | 0.04 |
ENST00000375629.3
ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS
|
isoleucyl-tRNA synthetase |
| chr8_+_30244580 | 0.03 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr12_+_26164645 | 0.03 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr4_-_36245561 | 0.03 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr12_-_25150409 | 0.03 |
ENST00000549262.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr9_-_95056010 | 0.03 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr2_-_190044480 | 0.03 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr2_+_161993465 | 0.03 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr12_-_71148357 | 0.03 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr10_+_90484301 | 0.03 |
ENST00000404190.1
|
LIPK
|
lipase, family member K |
| chrX_-_13835147 | 0.03 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr6_+_131958436 | 0.03 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr18_+_29027696 | 0.03 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr17_+_79849872 | 0.03 |
ENST00000584197.1
ENST00000583839.1 |
ANAPC11
|
anaphase promoting complex subunit 11 |
| chr17_+_67759813 | 0.03 |
ENST00000587241.1
|
AC003051.1
|
AC003051.1 |
| chr18_-_74839891 | 0.03 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr7_+_115862858 | 0.03 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr12_+_26348246 | 0.03 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr18_+_47087390 | 0.03 |
ENST00000583083.1
|
LIPG
|
lipase, endothelial |
| chr6_-_161695042 | 0.03 |
ENST00000366908.5
ENST00000366911.5 ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr11_+_12302492 | 0.03 |
ENST00000533534.1
|
MICALCL
|
MICAL C-terminal like |
| chr1_-_205091115 | 0.03 |
ENST00000264515.6
ENST00000367164.1 |
RBBP5
|
retinoblastoma binding protein 5 |
| chr8_-_91095099 | 0.03 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr17_+_74463650 | 0.03 |
ENST00000392492.3
|
AANAT
|
aralkylamine N-acetyltransferase |
| chr3_-_27498235 | 0.03 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr17_-_5321549 | 0.03 |
ENST00000572809.1
|
NUP88
|
nucleoporin 88kDa |
| chr11_-_117186946 | 0.03 |
ENST00000313005.6
ENST00000528053.1 |
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr17_+_8191815 | 0.03 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr8_+_76452097 | 0.03 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr1_-_160924589 | 0.03 |
ENST00000368029.3
|
ITLN2
|
intelectin 2 |
| chr1_+_81771806 | 0.03 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr1_+_233086326 | 0.03 |
ENST00000366628.5
ENST00000366627.4 |
NTPCR
|
nucleoside-triphosphatase, cancer-related |
| chr15_+_75970672 | 0.03 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr15_-_65321943 | 0.03 |
ENST00000220058.4
|
MTFMT
|
mitochondrial methionyl-tRNA formyltransferase |
| chr10_+_51572408 | 0.03 |
ENST00000374082.1
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr4_+_186990298 | 0.03 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr17_+_1674982 | 0.03 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr5_+_81575281 | 0.03 |
ENST00000380167.4
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU1F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.3 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.4 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.3 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.3 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0070839 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) divalent metal ion export(GO:0070839) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.0 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.4 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.2 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.0 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |