Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
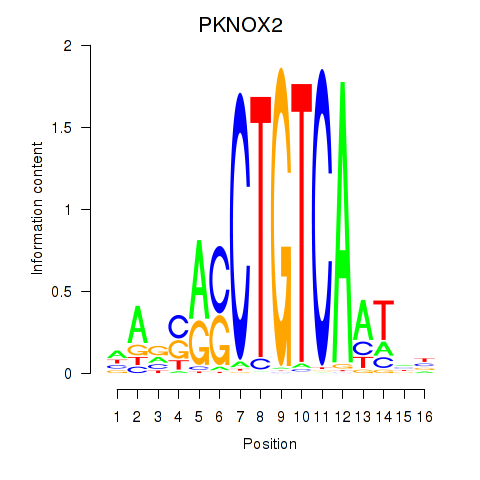
Results for PKNOX2
Z-value: 0.76
Transcription factors associated with PKNOX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PKNOX2
|
ENSG00000165495.11 | PBX/knotted 1 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PKNOX2 | hg19_v2_chr11_+_125034586_125034604 | 0.65 | 1.6e-01 | Click! |
Activity profile of PKNOX2 motif
Sorted Z-values of PKNOX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_99837856 | 0.27 |
ENST00000518165.1
ENST00000419617.2 |
STK3
|
serine/threonine kinase 3 |
| chr2_+_113321939 | 0.26 |
ENST00000458012.2
|
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr5_+_72509751 | 0.25 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr16_+_2880157 | 0.24 |
ENST00000382280.3
|
ZG16B
|
zymogen granule protein 16B |
| chr19_+_41281060 | 0.23 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr18_+_3449330 | 0.22 |
ENST00000549253.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr15_+_96904487 | 0.22 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr19_+_57050317 | 0.22 |
ENST00000301318.3
ENST00000591844.1 |
ZFP28
|
ZFP28 zinc finger protein |
| chr13_+_20268547 | 0.22 |
ENST00000601204.1
|
AL354808.2
|
AL354808.2 |
| chr10_+_94608218 | 0.22 |
ENST00000371543.1
|
EXOC6
|
exocyst complex component 6 |
| chr6_+_17600576 | 0.21 |
ENST00000259963.3
|
FAM8A1
|
family with sequence similarity 8, member A1 |
| chr2_-_202316169 | 0.20 |
ENST00000430254.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr12_+_113495492 | 0.20 |
ENST00000257600.3
|
DTX1
|
deltex homolog 1 (Drosophila) |
| chr3_-_52719888 | 0.19 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr5_-_132073111 | 0.19 |
ENST00000403231.1
|
KIF3A
|
kinesin family member 3A |
| chr18_-_52626622 | 0.18 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr16_+_2880369 | 0.18 |
ENST00000572863.1
|
ZG16B
|
zymogen granule protein 16B |
| chr11_+_74660278 | 0.18 |
ENST00000263672.6
ENST00000530257.1 ENST00000526361.1 ENST00000532972.1 |
SPCS2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr22_-_31885514 | 0.18 |
ENST00000397525.1
|
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr19_+_41281282 | 0.17 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chr2_+_202316392 | 0.16 |
ENST00000194530.3
ENST00000392249.2 |
STRADB
|
STE20-related kinase adaptor beta |
| chr6_+_111195973 | 0.16 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr9_-_123639304 | 0.16 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr14_+_61995722 | 0.16 |
ENST00000556347.1
|
RP11-47I22.4
|
RP11-47I22.4 |
| chr5_-_81046841 | 0.16 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr4_-_157892167 | 0.16 |
ENST00000541126.1
|
PDGFC
|
platelet derived growth factor C |
| chr3_+_52719936 | 0.15 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr2_+_196440692 | 0.15 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr13_-_48575376 | 0.15 |
ENST00000434484.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr17_-_29151794 | 0.15 |
ENST00000324238.6
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr8_+_57124245 | 0.15 |
ENST00000521831.1
ENST00000355315.3 ENST00000303759.3 ENST00000517636.1 ENST00000517933.1 ENST00000518801.1 ENST00000523975.1 ENST00000396723.5 ENST00000523061.1 ENST00000521524.1 |
CHCHD7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr3_-_101395936 | 0.15 |
ENST00000461821.1
|
ZBTB11
|
zinc finger and BTB domain containing 11 |
| chr4_-_157892055 | 0.15 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr6_+_41021027 | 0.15 |
ENST00000244669.2
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr5_-_139283982 | 0.14 |
ENST00000340391.3
|
NRG2
|
neuregulin 2 |
| chr3_+_186692745 | 0.14 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr9_+_6758109 | 0.14 |
ENST00000536108.1
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr8_+_11666649 | 0.13 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr11_-_111741994 | 0.13 |
ENST00000398006.2
|
ALG9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr2_+_191208196 | 0.13 |
ENST00000392329.2
ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr5_+_125758865 | 0.13 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr17_-_29151686 | 0.13 |
ENST00000544695.1
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr6_+_150070831 | 0.13 |
ENST00000367380.5
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr16_-_1275257 | 0.12 |
ENST00000234798.4
|
TPSG1
|
tryptase gamma 1 |
| chr15_+_68346501 | 0.12 |
ENST00000249636.6
|
PIAS1
|
protein inhibitor of activated STAT, 1 |
| chr17_-_27405875 | 0.12 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr12_+_62654155 | 0.11 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr10_+_28822636 | 0.11 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr5_+_125758813 | 0.11 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr17_+_8316442 | 0.11 |
ENST00000582812.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr2_+_64681219 | 0.11 |
ENST00000238875.5
|
LGALSL
|
lectin, galactoside-binding-like |
| chr2_-_242842587 | 0.11 |
ENST00000404031.1
|
AC131097.4
|
Protein LOC285095 |
| chr3_-_33481835 | 0.11 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr14_-_54421190 | 0.11 |
ENST00000417573.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr6_-_101329191 | 0.10 |
ENST00000324723.6
ENST00000369162.2 ENST00000522650.1 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr2_+_191208601 | 0.10 |
ENST00000413239.1
ENST00000431594.1 ENST00000444194.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr6_-_101329157 | 0.10 |
ENST00000369143.2
|
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr6_+_150070857 | 0.10 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chrX_-_80377162 | 0.10 |
ENST00000430960.1
ENST00000447319.1 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr6_+_138188378 | 0.09 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr18_-_67872891 | 0.09 |
ENST00000454359.1
ENST00000437017.1 |
RTTN
|
rotatin |
| chr16_+_2880254 | 0.09 |
ENST00000570670.1
|
ZG16B
|
zymogen granule protein 16B |
| chr15_+_59908633 | 0.09 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr18_-_54318353 | 0.09 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr3_+_113667354 | 0.09 |
ENST00000491556.1
|
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chrX_-_128788914 | 0.09 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr3_-_52719810 | 0.09 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr9_-_99637820 | 0.08 |
ENST00000289032.8
ENST00000535338.1 |
ZNF782
|
zinc finger protein 782 |
| chr3_-_33482002 | 0.08 |
ENST00000283628.5
ENST00000456378.1 |
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chrX_+_16804544 | 0.08 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr22_+_17082732 | 0.08 |
ENST00000558085.2
ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1
|
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr14_+_24600484 | 0.08 |
ENST00000267426.5
|
FITM1
|
fat storage-inducing transmembrane protein 1 |
| chr18_-_54305658 | 0.08 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chr2_+_64681103 | 0.08 |
ENST00000464281.1
|
LGALSL
|
lectin, galactoside-binding-like |
| chr9_+_125027127 | 0.08 |
ENST00000441707.1
ENST00000373723.5 ENST00000373729.1 |
MRRF
|
mitochondrial ribosome recycling factor |
| chr4_-_157892498 | 0.08 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr9_-_132805430 | 0.08 |
ENST00000446176.2
ENST00000355681.3 ENST00000420781.1 |
FNBP1
|
formin binding protein 1 |
| chr5_-_81046904 | 0.08 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr5_-_81046922 | 0.07 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr9_+_133454943 | 0.07 |
ENST00000319725.9
|
FUBP3
|
far upstream element (FUSE) binding protein 3 |
| chr12_+_69201923 | 0.07 |
ENST00000462284.1
ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr6_+_37225540 | 0.07 |
ENST00000373491.3
|
TBC1D22B
|
TBC1 domain family, member 22B |
| chr11_-_132813627 | 0.07 |
ENST00000374778.4
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr20_+_43514320 | 0.07 |
ENST00000372839.3
ENST00000428262.1 ENST00000445830.1 |
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta |
| chrX_-_80377118 | 0.07 |
ENST00000373250.3
|
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr2_-_202316260 | 0.07 |
ENST00000332624.3
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr3_+_32147997 | 0.07 |
ENST00000282541.5
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr11_-_46867780 | 0.07 |
ENST00000529230.1
ENST00000415402.1 ENST00000312055.5 |
CKAP5
|
cytoskeleton associated protein 5 |
| chr2_-_18770812 | 0.06 |
ENST00000359846.2
ENST00000304081.4 ENST00000600945.1 ENST00000532967.1 ENST00000444297.2 |
NT5C1B
NT5C1B-RDH14
|
5'-nucleotidase, cytosolic IB NT5C1B-RDH14 readthrough |
| chr8_+_133787586 | 0.06 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr18_+_3449821 | 0.06 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr6_-_37225391 | 0.06 |
ENST00000356757.2
|
TMEM217
|
transmembrane protein 217 |
| chr11_-_71810258 | 0.06 |
ENST00000544594.1
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr5_+_118406796 | 0.06 |
ENST00000503802.1
|
DMXL1
|
Dmx-like 1 |
| chr4_-_83483094 | 0.06 |
ENST00000508701.1
ENST00000454948.3 |
TMEM150C
|
transmembrane protein 150C |
| chr2_-_190446738 | 0.06 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr16_+_57481382 | 0.06 |
ENST00000564655.1
ENST00000567072.1 ENST00000567933.1 ENST00000563166.1 |
COQ9
|
coenzyme Q9 |
| chr1_-_179112189 | 0.06 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr16_-_57481278 | 0.06 |
ENST00000567751.1
ENST00000568940.1 ENST00000563341.1 ENST00000565961.1 ENST00000569370.1 ENST00000567518.1 ENST00000565786.1 ENST00000394391.4 |
CIAPIN1
|
cytokine induced apoptosis inhibitor 1 |
| chr9_+_6758024 | 0.05 |
ENST00000442236.2
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr11_-_119247004 | 0.05 |
ENST00000531070.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr1_+_66258846 | 0.05 |
ENST00000341517.4
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr18_+_3450161 | 0.05 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr1_-_35658736 | 0.05 |
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich |
| chr1_+_110577229 | 0.05 |
ENST00000369795.3
ENST00000369794.2 |
STRIP1
|
striatin interacting protein 1 |
| chr16_-_30457048 | 0.05 |
ENST00000500504.2
ENST00000542752.1 |
SEPHS2
|
selenophosphate synthetase 2 |
| chr18_+_54318616 | 0.05 |
ENST00000254442.3
|
WDR7
|
WD repeat domain 7 |
| chr10_+_70661014 | 0.05 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr8_+_72755796 | 0.05 |
ENST00000524152.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr12_+_62654119 | 0.05 |
ENST00000353364.3
ENST00000549523.1 ENST00000280377.5 |
USP15
|
ubiquitin specific peptidase 15 |
| chr2_-_61765732 | 0.05 |
ENST00000443240.1
ENST00000436018.1 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr11_+_120207787 | 0.05 |
ENST00000397843.2
ENST00000356641.3 |
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr1_+_113263199 | 0.05 |
ENST00000361886.3
|
FAM19A3
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A3 |
| chr16_+_2880296 | 0.05 |
ENST00000571723.1
|
ZG16B
|
zymogen granule protein 16B |
| chr14_-_21994337 | 0.05 |
ENST00000537235.1
ENST00000450879.2 |
SALL2
|
spalt-like transcription factor 2 |
| chrX_-_77225135 | 0.05 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr19_+_41281416 | 0.04 |
ENST00000597140.1
|
MIA
|
melanoma inhibitory activity |
| chr9_-_123639600 | 0.04 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr5_+_118407053 | 0.04 |
ENST00000311085.8
ENST00000539542.1 |
DMXL1
|
Dmx-like 1 |
| chr7_-_156803329 | 0.04 |
ENST00000252971.6
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr4_+_169418255 | 0.04 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr17_-_40337470 | 0.04 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr2_-_61765315 | 0.04 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr21_-_32931290 | 0.04 |
ENST00000286827.3
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1 |
| chr3_+_185080908 | 0.04 |
ENST00000265026.3
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr1_-_201081579 | 0.04 |
ENST00000367338.3
ENST00000362061.3 |
CACNA1S
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr2_+_219524473 | 0.04 |
ENST00000439945.1
ENST00000431802.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr11_-_132812987 | 0.04 |
ENST00000541867.1
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr1_+_161736072 | 0.04 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr8_-_37594944 | 0.04 |
ENST00000330539.1
|
RP11-863K10.7
|
Uncharacterized protein |
| chr8_-_20161466 | 0.03 |
ENST00000381569.1
|
LZTS1
|
leucine zipper, putative tumor suppressor 1 |
| chr4_-_46391367 | 0.03 |
ENST00000503806.1
ENST00000356504.1 ENST00000514090.1 ENST00000506961.1 |
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr15_-_37392086 | 0.03 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr17_-_35969409 | 0.03 |
ENST00000394378.2
ENST00000585472.1 ENST00000591288.1 ENST00000502449.2 ENST00000345615.4 ENST00000346661.4 ENST00000585689.1 ENST00000339208.6 |
SYNRG
|
synergin, gamma |
| chr5_-_52405564 | 0.03 |
ENST00000510818.2
ENST00000396954.3 ENST00000508922.1 ENST00000361377.4 ENST00000582677.1 ENST00000584946.1 ENST00000450852.3 |
MOCS2
|
molybdenum cofactor synthesis 2 |
| chr2_-_74780176 | 0.03 |
ENST00000409549.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr3_+_183899770 | 0.03 |
ENST00000442686.1
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr4_+_128703295 | 0.03 |
ENST00000296464.4
ENST00000508549.1 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr5_-_94620239 | 0.03 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr10_+_49514698 | 0.03 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr3_-_52719912 | 0.03 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chr11_+_129245796 | 0.02 |
ENST00000281437.4
|
BARX2
|
BARX homeobox 2 |
| chr10_-_101945771 | 0.02 |
ENST00000370408.2
ENST00000407654.3 |
ERLIN1
|
ER lipid raft associated 1 |
| chr4_+_3076388 | 0.02 |
ENST00000355072.5
|
HTT
|
huntingtin |
| chr3_+_23244780 | 0.02 |
ENST00000396703.1
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr10_+_51565188 | 0.02 |
ENST00000430396.2
ENST00000374087.4 ENST00000414907.2 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr7_-_148580563 | 0.02 |
ENST00000476773.1
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr4_+_140222609 | 0.02 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr9_+_87285539 | 0.02 |
ENST00000359847.3
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr8_+_31496809 | 0.02 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr1_-_234667504 | 0.02 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr10_-_124768300 | 0.01 |
ENST00000368886.5
|
IKZF5
|
IKAROS family zinc finger 5 (Pegasus) |
| chr7_-_104909435 | 0.01 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr2_+_128848740 | 0.01 |
ENST00000375990.3
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr16_+_57481349 | 0.01 |
ENST00000262507.6
ENST00000565964.1 |
COQ9
|
coenzyme Q9 |
| chr5_+_125759140 | 0.01 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr17_-_26220366 | 0.01 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr2_+_113033164 | 0.01 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr4_+_166300084 | 0.01 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr11_-_59383617 | 0.01 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr9_+_128510454 | 0.01 |
ENST00000491787.3
ENST00000447726.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr2_+_128848881 | 0.01 |
ENST00000259253.6
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr12_+_49621658 | 0.01 |
ENST00000541364.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr4_-_184241927 | 0.01 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr18_+_54318566 | 0.00 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr6_+_42123141 | 0.00 |
ENST00000418175.1
ENST00000541991.1 ENST00000053469.4 ENST00000394237.1 ENST00000372963.1 |
GUCA1A
RP1-139D8.6
|
guanylate cyclase activator 1A (retina) RP1-139D8.6 |
| chr1_-_100598444 | 0.00 |
ENST00000535161.1
ENST00000287482.5 |
SASS6
|
spindle assembly 6 homolog (C. elegans) |
| chr11_-_8832521 | 0.00 |
ENST00000530438.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr4_-_149365827 | 0.00 |
ENST00000344721.4
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr22_+_30279144 | 0.00 |
ENST00000401950.2
ENST00000333027.3 ENST00000445401.1 ENST00000323630.5 ENST00000351488.3 |
MTMR3
|
myotubularin related protein 3 |
| chr16_+_50776021 | 0.00 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr9_-_73477826 | 0.00 |
ENST00000396285.1
ENST00000396292.4 ENST00000396280.5 ENST00000358082.3 ENST00000408909.2 ENST00000377097.3 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chrX_+_2924654 | 0.00 |
ENST00000381130.2
|
ARSH
|
arylsulfatase family, member H |
| chr2_-_219524193 | 0.00 |
ENST00000450560.1
ENST00000449707.1 ENST00000432460.1 ENST00000411696.2 |
ZNF142
|
zinc finger protein 142 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PKNOX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.2 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.2 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.1 | GO:0034148 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of osteoclast proliferation(GO:0090291) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.0 | 0.1 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.1 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.1 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0072192 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:0070839 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) divalent metal ion export(GO:0070839) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.0 | GO:1903979 | negative regulation of microglial cell activation(GO:1903979) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.0 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.2 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.1 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.2 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.0 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |