Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
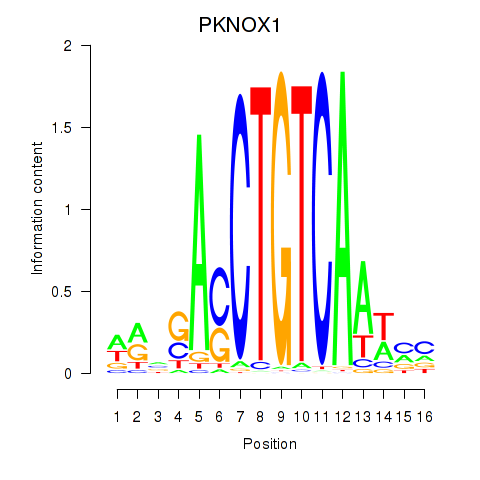
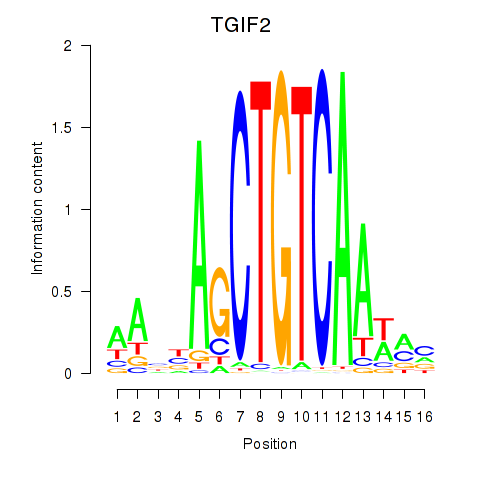
Results for PKNOX1_TGIF2
Z-value: 0.74
Transcription factors associated with PKNOX1_TGIF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PKNOX1
|
ENSG00000160199.10 | PBX/knotted 1 homeobox 1 |
|
TGIF2
|
ENSG00000118707.5 | TGFB induced factor homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TGIF2 | hg19_v2_chr20_+_35202909_35203075 | -0.85 | 3.3e-02 | Click! |
| PKNOX1 | hg19_v2_chr21_+_44394620_44394737 | 0.23 | 6.7e-01 | Click! |
Activity profile of PKNOX1_TGIF2 motif
Sorted Z-values of PKNOX1_TGIF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_191208601 | 0.82 |
ENST00000413239.1
ENST00000431594.1 ENST00000444194.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr1_-_63988846 | 0.60 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr18_-_54318353 | 0.53 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr9_-_33447584 | 0.52 |
ENST00000297991.4
|
AQP3
|
aquaporin 3 (Gill blood group) |
| chr4_-_175443484 | 0.52 |
ENST00000514584.1
ENST00000542498.1 ENST00000296521.7 ENST00000422112.2 ENST00000504433.1 |
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr15_+_96904487 | 0.50 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr1_+_63989004 | 0.48 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr5_+_150406527 | 0.47 |
ENST00000520059.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr4_-_175443943 | 0.45 |
ENST00000296522.6
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr7_+_79765071 | 0.41 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr16_-_79634595 | 0.41 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr2_+_29353520 | 0.39 |
ENST00000438819.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr3_+_186692745 | 0.38 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr6_+_26124373 | 0.37 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr17_-_77967433 | 0.35 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr8_-_99837856 | 0.34 |
ENST00000518165.1
ENST00000419617.2 |
STK3
|
serine/threonine kinase 3 |
| chr19_+_39759154 | 0.34 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr22_+_20905422 | 0.31 |
ENST00000424287.1
ENST00000423862.1 |
MED15
|
mediator complex subunit 15 |
| chr7_+_142448053 | 0.28 |
ENST00000422143.2
|
TRBV29-1
|
T cell receptor beta variable 29-1 |
| chr2_-_192015697 | 0.27 |
ENST00000409995.1
|
STAT4
|
signal transducer and activator of transcription 4 |
| chr1_-_145715565 | 0.26 |
ENST00000369288.2
ENST00000369290.1 ENST00000401557.3 |
CD160
|
CD160 molecule |
| chr18_+_3449330 | 0.25 |
ENST00000549253.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr11_+_827553 | 0.24 |
ENST00000528542.2
ENST00000450448.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr12_+_69004619 | 0.24 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr18_-_54305658 | 0.23 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chr1_-_32384693 | 0.23 |
ENST00000602683.1
ENST00000470404.1 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr1_-_70671216 | 0.22 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr2_+_191208656 | 0.22 |
ENST00000458647.1
|
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr4_-_83483094 | 0.22 |
ENST00000508701.1
ENST00000454948.3 |
TMEM150C
|
transmembrane protein 150C |
| chr6_-_41673552 | 0.22 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr7_+_23637118 | 0.21 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr9_-_99180597 | 0.21 |
ENST00000375256.4
|
ZNF367
|
zinc finger protein 367 |
| chr2_-_61765315 | 0.21 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr3_+_38017264 | 0.21 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr4_+_166300084 | 0.20 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr18_-_52626622 | 0.20 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr16_-_30457048 | 0.19 |
ENST00000500504.2
ENST00000542752.1 |
SEPHS2
|
selenophosphate synthetase 2 |
| chr2_+_191208791 | 0.19 |
ENST00000423767.1
ENST00000451089.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr14_+_61995722 | 0.19 |
ENST00000556347.1
|
RP11-47I22.4
|
RP11-47I22.4 |
| chr2_+_223725723 | 0.19 |
ENST00000535678.1
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr5_-_42887494 | 0.19 |
ENST00000514218.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr1_-_205391178 | 0.19 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr18_+_54318893 | 0.19 |
ENST00000593058.1
|
WDR7
|
WD repeat domain 7 |
| chr22_+_20905269 | 0.19 |
ENST00000457322.1
|
MED15
|
mediator complex subunit 15 |
| chr5_-_55008072 | 0.19 |
ENST00000512208.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr9_-_134615326 | 0.18 |
ENST00000438647.1
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr6_-_35656685 | 0.18 |
ENST00000539068.1
ENST00000540787.1 |
FKBP5
|
FK506 binding protein 5 |
| chr7_+_23637763 | 0.18 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr6_+_111195973 | 0.18 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr15_+_51200871 | 0.17 |
ENST00000560508.1
|
AP4E1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr2_-_190446738 | 0.17 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr7_+_39605966 | 0.17 |
ENST00000223273.2
ENST00000448268.1 ENST00000432096.2 |
YAE1D1
|
Yae1 domain containing 1 |
| chr9_-_100459639 | 0.17 |
ENST00000375128.4
|
XPA
|
xeroderma pigmentosum, complementation group A |
| chr17_+_26833250 | 0.17 |
ENST00000577936.1
ENST00000579795.1 |
FOXN1
|
forkhead box N1 |
| chr13_+_20268547 | 0.17 |
ENST00000601204.1
|
AL354808.2
|
AL354808.2 |
| chr2_-_102003987 | 0.16 |
ENST00000324768.5
|
CREG2
|
cellular repressor of E1A-stimulated genes 2 |
| chr5_+_43603229 | 0.16 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr13_-_41837620 | 0.16 |
ENST00000379477.1
ENST00000452359.1 ENST00000379480.4 ENST00000430347.2 |
MTRF1
|
mitochondrial translational release factor 1 |
| chr8_-_17555164 | 0.16 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr5_-_132073111 | 0.16 |
ENST00000403231.1
|
KIF3A
|
kinesin family member 3A |
| chr10_-_118764862 | 0.16 |
ENST00000260777.10
|
KIAA1598
|
KIAA1598 |
| chr1_-_247171347 | 0.16 |
ENST00000339986.7
ENST00000487338.2 |
ZNF695
|
zinc finger protein 695 |
| chr3_-_142166904 | 0.16 |
ENST00000264951.4
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr12_+_69201923 | 0.15 |
ENST00000462284.1
ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr11_-_1782625 | 0.15 |
ENST00000438213.1
|
CTSD
|
cathepsin D |
| chr6_+_53794780 | 0.15 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chr2_+_10560147 | 0.15 |
ENST00000422133.1
|
HPCAL1
|
hippocalcin-like 1 |
| chr8_+_97657449 | 0.15 |
ENST00000220763.5
|
CPQ
|
carboxypeptidase Q |
| chr14_+_101295948 | 0.15 |
ENST00000452514.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr19_+_11877838 | 0.15 |
ENST00000357901.4
ENST00000454339.2 |
ZNF441
|
zinc finger protein 441 |
| chr10_-_4720333 | 0.15 |
ENST00000430998.2
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr3_-_142166796 | 0.15 |
ENST00000392981.2
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr4_+_128886584 | 0.15 |
ENST00000513371.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr11_+_27062272 | 0.15 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr4_-_47839966 | 0.14 |
ENST00000273857.4
ENST00000505909.1 ENST00000502252.1 |
CORIN
|
corin, serine peptidase |
| chr2_+_198380289 | 0.14 |
ENST00000233892.4
ENST00000409916.1 |
MOB4
|
MOB family member 4, phocein |
| chr11_+_6411636 | 0.14 |
ENST00000299397.3
ENST00000356761.2 ENST00000342245.4 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr4_+_86396321 | 0.14 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr10_+_94608245 | 0.14 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr2_+_191208196 | 0.14 |
ENST00000392329.2
ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr2_-_61765732 | 0.14 |
ENST00000443240.1
ENST00000436018.1 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr6_-_101329191 | 0.14 |
ENST00000324723.6
ENST00000369162.2 ENST00000522650.1 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr3_-_3221358 | 0.14 |
ENST00000424814.1
ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN
|
cereblon |
| chr20_-_36156264 | 0.14 |
ENST00000445723.1
ENST00000414080.1 |
BLCAP
|
bladder cancer associated protein |
| chr3_-_149093499 | 0.13 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr15_+_49913201 | 0.13 |
ENST00000329873.5
ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1
|
DTW domain containing 1 |
| chr3_-_143567262 | 0.13 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr11_+_6411670 | 0.13 |
ENST00000530395.1
ENST00000527275.1 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr5_-_81046904 | 0.13 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr9_+_123884038 | 0.13 |
ENST00000373847.1
|
CNTRL
|
centriolin |
| chr4_+_8183793 | 0.13 |
ENST00000509119.1
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr2_+_233925064 | 0.13 |
ENST00000359570.5
ENST00000538935.1 |
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr6_+_41021027 | 0.13 |
ENST00000244669.2
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr15_+_49913175 | 0.12 |
ENST00000403028.3
|
DTWD1
|
DTW domain containing 1 |
| chr12_+_62654155 | 0.12 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr15_-_31283798 | 0.12 |
ENST00000435680.1
ENST00000425768.1 |
MTMR10
|
myotubularin related protein 10 |
| chr12_+_34175398 | 0.12 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr8_-_8318847 | 0.12 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr19_-_11849697 | 0.12 |
ENST00000586121.1
ENST00000431998.1 ENST00000341191.6 ENST00000545749.1 ENST00000440527.1 |
ZNF823
|
zinc finger protein 823 |
| chr5_-_81046841 | 0.12 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr6_-_119031228 | 0.12 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr9_-_123639600 | 0.11 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chrX_-_101397433 | 0.11 |
ENST00000372774.3
|
TCEAL6
|
transcription elongation factor A (SII)-like 6 |
| chr4_-_114682224 | 0.11 |
ENST00000342666.5
ENST00000515496.1 ENST00000514328.1 ENST00000508738.1 ENST00000379773.2 |
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr13_+_73302047 | 0.11 |
ENST00000377814.2
ENST00000377815.3 ENST00000390667.5 |
BORA
|
bora, aurora kinase A activator |
| chr11_+_64052294 | 0.11 |
ENST00000536667.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr16_+_2588012 | 0.11 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr12_+_97306295 | 0.11 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr7_-_156685841 | 0.11 |
ENST00000354505.4
ENST00000540390.1 |
LMBR1
|
limb development membrane protein 1 |
| chr17_+_13972807 | 0.11 |
ENST00000429152.2
ENST00000261643.3 ENST00000536205.1 ENST00000537334.1 |
COX10
|
cytochrome c oxidase assembly homolog 10 (yeast) |
| chr3_-_142166846 | 0.11 |
ENST00000463916.1
ENST00000544157.1 |
XRN1
|
5'-3' exoribonuclease 1 |
| chr14_-_92198403 | 0.11 |
ENST00000553329.1
ENST00000256343.3 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr9_-_86432547 | 0.11 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr3_+_111260980 | 0.11 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr22_-_50219548 | 0.11 |
ENST00000404034.1
|
BRD1
|
bromodomain containing 1 |
| chr15_+_59908633 | 0.10 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr19_+_36393422 | 0.10 |
ENST00000437550.2
|
HCST
|
hematopoietic cell signal transducer |
| chr11_+_27062502 | 0.10 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr21_+_38792602 | 0.10 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr11_-_67415048 | 0.10 |
ENST00000529256.1
|
ACY3
|
aspartoacylase (aminocyclase) 3 |
| chr15_-_37391614 | 0.10 |
ENST00000219869.9
|
MEIS2
|
Meis homeobox 2 |
| chr22_-_37172111 | 0.10 |
ENST00000417951.2
ENST00000430701.1 ENST00000433985.2 |
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr6_-_4347271 | 0.10 |
ENST00000437430.2
|
RP3-527G5.1
|
RP3-527G5.1 |
| chr4_-_114682364 | 0.10 |
ENST00000511664.1
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr7_+_99971129 | 0.10 |
ENST00000394000.2
ENST00000350573.2 |
PILRA
|
paired immunoglobin-like type 2 receptor alpha |
| chr19_-_12551849 | 0.10 |
ENST00000595562.1
ENST00000301547.5 |
CTD-3105H18.16
ZNF443
|
Uncharacterized protein zinc finger protein 443 |
| chr19_+_52800410 | 0.10 |
ENST00000595962.1
ENST00000598016.1 ENST00000334564.7 ENST00000490272.1 |
ZNF480
|
zinc finger protein 480 |
| chr21_-_34144157 | 0.10 |
ENST00000331923.4
|
PAXBP1
|
PAX3 and PAX7 binding protein 1 |
| chr7_+_70229899 | 0.09 |
ENST00000443672.1
|
AUTS2
|
autism susceptibility candidate 2 |
| chr1_+_77333117 | 0.09 |
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr4_+_159593418 | 0.09 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr12_+_11081828 | 0.09 |
ENST00000381847.3
ENST00000396400.3 |
PRH2
|
proline-rich protein HaeIII subfamily 2 |
| chr2_+_62900986 | 0.09 |
ENST00000405015.3
ENST00000413434.1 ENST00000426940.1 ENST00000449820.1 |
EHBP1
|
EH domain binding protein 1 |
| chr11_-_71810258 | 0.09 |
ENST00000544594.1
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr16_-_20817753 | 0.09 |
ENST00000389345.5
ENST00000300005.3 ENST00000357967.4 ENST00000569729.1 |
ERI2
|
ERI1 exoribonuclease family member 2 |
| chr5_-_81046922 | 0.09 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr2_+_113321939 | 0.08 |
ENST00000458012.2
|
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr10_-_73848764 | 0.08 |
ENST00000317376.4
ENST00000412663.1 |
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr12_+_57522692 | 0.08 |
ENST00000554174.1
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr19_+_9296279 | 0.08 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr15_-_77197620 | 0.08 |
ENST00000565970.1
ENST00000563290.1 ENST00000565372.1 ENST00000564177.1 ENST00000568382.1 ENST00000563919.1 |
SCAPER
|
S-phase cyclin A-associated protein in the ER |
| chr17_-_71223839 | 0.08 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr10_+_28822636 | 0.08 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr12_-_11036844 | 0.08 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr7_-_148580563 | 0.08 |
ENST00000476773.1
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr5_-_55008101 | 0.08 |
ENST00000506624.1
ENST00000513275.1 ENST00000513993.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr1_+_186649754 | 0.08 |
ENST00000608917.1
|
RP5-973M2.2
|
RP5-973M2.2 |
| chr13_+_33160553 | 0.08 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr3_+_183899770 | 0.08 |
ENST00000442686.1
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr12_+_69186125 | 0.08 |
ENST00000399333.3
|
AC124890.1
|
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
| chr10_+_94608218 | 0.08 |
ENST00000371543.1
|
EXOC6
|
exocyst complex component 6 |
| chr15_+_36871983 | 0.08 |
ENST00000437989.2
ENST00000569302.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr12_-_75784669 | 0.08 |
ENST00000552497.1
ENST00000551829.1 ENST00000436898.1 ENST00000442339.2 |
CAPS2
|
calcyphosine 2 |
| chr1_+_222886694 | 0.08 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr8_+_38244638 | 0.08 |
ENST00000526356.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr2_-_183903133 | 0.08 |
ENST00000361354.4
|
NCKAP1
|
NCK-associated protein 1 |
| chr21_-_34143971 | 0.08 |
ENST00000290178.4
|
PAXBP1
|
PAX3 and PAX7 binding protein 1 |
| chr17_+_7621045 | 0.07 |
ENST00000570791.1
|
DNAH2
|
dynein, axonemal, heavy chain 2 |
| chr16_+_16484691 | 0.07 |
ENST00000344087.4
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr6_+_42123141 | 0.07 |
ENST00000418175.1
ENST00000541991.1 ENST00000053469.4 ENST00000394237.1 ENST00000372963.1 |
GUCA1A
RP1-139D8.6
|
guanylate cyclase activator 1A (retina) RP1-139D8.6 |
| chr16_+_2587965 | 0.07 |
ENST00000342085.4
ENST00000566659.1 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr4_+_88928777 | 0.07 |
ENST00000237596.2
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant) |
| chr2_+_175260451 | 0.07 |
ENST00000458563.1
ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3
|
secernin 3 |
| chr3_-_33481835 | 0.07 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr9_+_34652164 | 0.07 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr6_+_155334780 | 0.07 |
ENST00000538270.1
ENST00000535231.1 |
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr19_-_58951496 | 0.07 |
ENST00000254166.3
|
ZNF132
|
zinc finger protein 132 |
| chr15_-_37392086 | 0.07 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr10_+_121485588 | 0.07 |
ENST00000361976.2
ENST00000369083.3 |
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr10_+_114710516 | 0.07 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_+_57522439 | 0.07 |
ENST00000338962.4
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr5_+_118407053 | 0.07 |
ENST00000311085.8
ENST00000539542.1 |
DMXL1
|
Dmx-like 1 |
| chr2_-_37068530 | 0.07 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr1_-_203151933 | 0.07 |
ENST00000404436.2
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39) |
| chr7_+_99971068 | 0.06 |
ENST00000198536.2
ENST00000453419.1 |
PILRA
|
paired immunoglobin-like type 2 receptor alpha |
| chr6_-_28220002 | 0.06 |
ENST00000377294.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr18_+_39535239 | 0.06 |
ENST00000585528.1
|
PIK3C3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr6_-_131949200 | 0.06 |
ENST00000539158.1
ENST00000368058.1 |
MED23
|
mediator complex subunit 23 |
| chr10_-_124768300 | 0.06 |
ENST00000368886.5
|
IKZF5
|
IKAROS family zinc finger 5 (Pegasus) |
| chr13_+_24844857 | 0.06 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr7_+_94536514 | 0.06 |
ENST00000413325.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr16_+_29789561 | 0.06 |
ENST00000400752.4
|
ZG16
|
zymogen granule protein 16 |
| chr10_-_99052382 | 0.06 |
ENST00000453547.2
ENST00000316676.8 ENST00000358308.3 ENST00000466484.1 ENST00000358531.4 |
ARHGAP19-SLIT1
ARHGAP19
|
ARHGAP19-SLIT1 readthrough (NMD candidate) Rho GTPase activating protein 19 |
| chr5_-_52405564 | 0.06 |
ENST00000510818.2
ENST00000396954.3 ENST00000508922.1 ENST00000361377.4 ENST00000582677.1 ENST00000584946.1 ENST00000450852.3 |
MOCS2
|
molybdenum cofactor synthesis 2 |
| chr7_-_7575477 | 0.06 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr16_+_57139933 | 0.06 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chr1_-_247335269 | 0.06 |
ENST00000543802.2
ENST00000491356.1 ENST00000472531.1 ENST00000340684.6 |
ZNF124
|
zinc finger protein 124 |
| chr1_+_210406121 | 0.06 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr16_+_618837 | 0.06 |
ENST00000409439.2
|
PIGQ
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr10_-_118765081 | 0.06 |
ENST00000392903.2
ENST00000355371.4 |
KIAA1598
|
KIAA1598 |
| chr5_-_94620239 | 0.06 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr22_+_20905286 | 0.06 |
ENST00000428629.1
|
MED15
|
mediator complex subunit 15 |
| chr18_+_48918368 | 0.05 |
ENST00000583982.1
ENST00000578152.1 ENST00000583609.1 ENST00000435144.1 ENST00000580841.1 |
RP11-267C16.1
|
RP11-267C16.1 |
| chr5_-_55008136 | 0.05 |
ENST00000503891.1
ENST00000507109.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr12_+_85430110 | 0.05 |
ENST00000393212.3
ENST00000393217.2 |
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1 |
| chr7_-_91772263 | 0.05 |
ENST00000435873.1
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1 |
| chr22_+_50946645 | 0.05 |
ENST00000420993.2
ENST00000395698.3 ENST00000395701.3 ENST00000523045.1 ENST00000299821.11 |
NCAPH2
|
non-SMC condensin II complex, subunit H2 |
| chr18_-_46987000 | 0.05 |
ENST00000442713.2
ENST00000269445.6 |
DYM
|
dymeclin |
| chr4_+_146539415 | 0.05 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr8_-_18541603 | 0.05 |
ENST00000428502.2
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PKNOX1_TGIF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 0.5 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.4 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.2 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.1 | 0.5 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.2 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.2 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.1 | 0.2 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.1 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.1 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.2 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.4 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0033488 | cholesterol biosynthetic process via 24,25-dihydrolanosterol(GO:0033488) |
| 0.0 | 0.2 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.0 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.2 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.6 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.0 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.3 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.3 | 1.0 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.5 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.2 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.2 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.2 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.2 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 0.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.2 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0048763 | HLH domain binding(GO:0043398) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0008398 | sterol 14-demethylase activity(GO:0008398) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.0 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |