Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
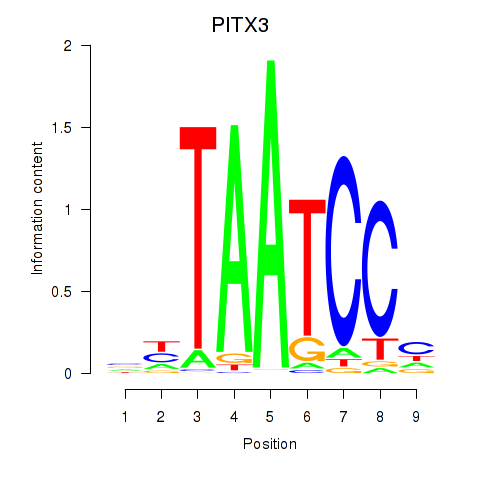
Results for PITX3
Z-value: 1.48
Transcription factors associated with PITX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PITX3
|
ENSG00000107859.5 | paired like homeodomain 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PITX3 | hg19_v2_chr10_-_104001231_104001274 | 0.21 | 7.0e-01 | Click! |
Activity profile of PITX3 motif
Sorted Z-values of PITX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_153013588 | 2.14 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chr15_-_80263506 | 1.13 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr12_+_22852791 | 0.99 |
ENST00000413794.2
|
RP11-114G22.1
|
RP11-114G22.1 |
| chr17_-_38859996 | 0.88 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr6_-_10694766 | 0.84 |
ENST00000460742.2
ENST00000259983.3 ENST00000379586.1 |
C6orf52
|
chromosome 6 open reading frame 52 |
| chr1_-_153363452 | 0.83 |
ENST00000368732.1
ENST00000368733.3 |
S100A8
|
S100 calcium binding protein A8 |
| chr1_-_153113927 | 0.79 |
ENST00000368752.4
|
SPRR2B
|
small proline-rich protein 2B |
| chr19_-_6690723 | 0.79 |
ENST00000601008.1
|
C3
|
complement component 3 |
| chr8_-_25281747 | 0.77 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr8_-_150563 | 0.72 |
ENST00000523795.2
|
RP11-585F1.10
|
Protein LOC100286914 |
| chr1_+_207262881 | 0.67 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr1_-_153029980 | 0.67 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr22_+_38609538 | 0.65 |
ENST00000407965.1
|
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr12_-_66317967 | 0.65 |
ENST00000601398.1
|
AC090673.2
|
Uncharacterized protein |
| chr12_+_7941989 | 0.64 |
ENST00000229307.4
|
NANOG
|
Nanog homeobox |
| chr1_-_161193349 | 0.63 |
ENST00000469730.2
ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2
|
apolipoprotein A-II |
| chr1_-_47655686 | 0.62 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr14_-_51863853 | 0.58 |
ENST00000556762.1
|
RP11-255G12.3
|
RP11-255G12.3 |
| chr20_+_55099542 | 0.56 |
ENST00000371328.3
|
FAM209A
|
family with sequence similarity 209, member A |
| chr1_-_153085984 | 0.55 |
ENST00000468739.1
|
SPRR2F
|
small proline-rich protein 2F |
| chr11_+_18287801 | 0.53 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr6_+_31895480 | 0.52 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr16_+_58010339 | 0.52 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr19_-_3772209 | 0.51 |
ENST00000555978.1
ENST00000555633.1 |
RAX2
|
retina and anterior neural fold homeobox 2 |
| chr11_+_18287721 | 0.51 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr18_+_61564389 | 0.50 |
ENST00000397996.2
ENST00000418725.1 |
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr3_+_172361483 | 0.49 |
ENST00000598405.1
|
AC007919.2
|
HCG1787166; PRO1163; Uncharacterized protein |
| chr17_+_41158742 | 0.48 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr4_+_154622652 | 0.48 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr9_+_74920408 | 0.47 |
ENST00000451152.1
|
RP11-63P12.6
|
RP11-63P12.6 |
| chr12_+_72058130 | 0.47 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr6_+_77484663 | 0.47 |
ENST00000607287.1
|
RP11-354K4.2
|
RP11-354K4.2 |
| chr7_+_16828866 | 0.46 |
ENST00000597084.1
|
AC073333.1
|
Uncharacterized protein |
| chr5_+_151338426 | 0.45 |
ENST00000524034.1
|
CTB-12O2.1
|
CTB-12O2.1 |
| chr10_-_51958906 | 0.45 |
ENST00000489640.1
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr18_+_68002675 | 0.43 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr12_+_113416340 | 0.43 |
ENST00000552756.1
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chrX_-_46759138 | 0.42 |
ENST00000377879.3
|
CXorf31
|
chromosome X open reading frame 31 |
| chr1_-_153044083 | 0.42 |
ENST00000341611.2
|
SPRR2B
|
small proline-rich protein 2B |
| chr5_+_142286887 | 0.42 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr6_+_12008986 | 0.42 |
ENST00000491710.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr4_-_165898768 | 0.41 |
ENST00000329314.5
|
TRIM61
|
tripartite motif containing 61 |
| chr19_+_36027660 | 0.41 |
ENST00000585510.1
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr5_-_150467221 | 0.40 |
ENST00000522226.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr3_-_49837254 | 0.40 |
ENST00000412678.2
ENST00000343366.4 ENST00000487256.1 |
CDHR4
|
cadherin-related family member 4 |
| chr16_+_84801852 | 0.40 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr14_-_31889737 | 0.39 |
ENST00000382464.2
|
HEATR5A
|
HEAT repeat containing 5A |
| chr15_+_45722727 | 0.38 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr16_-_57935277 | 0.38 |
ENST00000565942.1
|
CNGB1
|
cyclic nucleotide gated channel beta 1 |
| chrX_+_69488174 | 0.37 |
ENST00000480877.2
ENST00000307959.8 |
ARR3
|
arrestin 3, retinal (X-arrestin) |
| chr12_-_110883346 | 0.37 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr16_+_67195004 | 0.36 |
ENST00000523893.1
|
FBXL8
|
F-box and leucine-rich repeat protein 8 |
| chr19_+_58694742 | 0.36 |
ENST00000597528.1
ENST00000594839.1 |
ZNF274
|
zinc finger protein 274 |
| chr14_+_103592636 | 0.36 |
ENST00000333007.1
|
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr17_-_47925379 | 0.35 |
ENST00000352793.2
ENST00000334568.4 ENST00000398154.1 ENST00000436235.1 ENST00000326219.5 |
TAC4
|
tachykinin 4 (hemokinin) |
| chrX_+_71401570 | 0.35 |
ENST00000496835.2
ENST00000446576.1 |
PIN4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr11_-_105010320 | 0.35 |
ENST00000532895.1
ENST00000530950.1 |
CARD18
|
caspase recruitment domain family, member 18 |
| chr8_-_73793975 | 0.35 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr9_-_112970436 | 0.34 |
ENST00000400613.4
|
C9orf152
|
chromosome 9 open reading frame 152 |
| chr3_+_188664988 | 0.34 |
ENST00000433971.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr8_-_142011244 | 0.34 |
ENST00000340930.3
ENST00000520828.1 ENST00000524257.1 ENST00000523679.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr9_+_116343192 | 0.34 |
ENST00000471324.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr5_+_67485704 | 0.34 |
ENST00000520762.1
|
RP11-404L6.2
|
Uncharacterized protein |
| chr16_+_2016821 | 0.34 |
ENST00000569210.2
ENST00000569714.1 |
RNF151
|
ring finger protein 151 |
| chr6_-_133079022 | 0.33 |
ENST00000525289.1
ENST00000326499.6 |
VNN2
|
vanin 2 |
| chr1_+_169079823 | 0.33 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr4_-_467892 | 0.33 |
ENST00000506646.1
ENST00000505900.1 |
ZNF721
|
zinc finger protein 721 |
| chr12_+_113416191 | 0.33 |
ENST00000342315.4
ENST00000392583.2 |
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr17_-_48207115 | 0.33 |
ENST00000511964.1
|
SAMD14
|
sterile alpha motif domain containing 14 |
| chr12_-_2027639 | 0.33 |
ENST00000586184.1
ENST00000587995.1 ENST00000585732.1 |
CACNA2D4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr4_+_70916119 | 0.32 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chrX_-_101410762 | 0.32 |
ENST00000543160.1
ENST00000333643.3 |
BEX5
|
brain expressed, X-linked 5 |
| chr17_+_45908974 | 0.32 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr15_-_90039805 | 0.31 |
ENST00000544600.1
ENST00000268122.4 |
RHCG
|
Rh family, C glycoprotein |
| chr2_-_69664549 | 0.31 |
ENST00000450796.2
ENST00000484177.1 |
NFU1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr10_-_33405600 | 0.31 |
ENST00000414308.1
|
RP11-342D11.3
|
RP11-342D11.3 |
| chr5_+_36606700 | 0.31 |
ENST00000416645.2
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr4_+_81256871 | 0.30 |
ENST00000358105.3
ENST00000508675.1 |
C4orf22
|
chromosome 4 open reading frame 22 |
| chr9_+_75766763 | 0.30 |
ENST00000456643.1
ENST00000415424.1 |
ANXA1
|
annexin A1 |
| chr7_-_143956815 | 0.30 |
ENST00000493325.1
|
OR2A7
|
olfactory receptor, family 2, subfamily A, member 7 |
| chr2_-_68547061 | 0.30 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr11_-_321340 | 0.30 |
ENST00000526811.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr1_-_205391178 | 0.30 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr1_+_50459990 | 0.29 |
ENST00000448346.1
|
AL645730.2
|
AL645730.2 |
| chr1_-_150849047 | 0.29 |
ENST00000354396.2
ENST00000505755.1 |
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr19_-_17571722 | 0.29 |
ENST00000301944.2
|
NXNL1
|
nucleoredoxin-like 1 |
| chr11_+_125461607 | 0.29 |
ENST00000527606.1
|
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr12_+_104682667 | 0.28 |
ENST00000527335.1
|
TXNRD1
|
thioredoxin reductase 1 |
| chr4_-_87374330 | 0.28 |
ENST00000511328.1
ENST00000503911.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr19_-_17186229 | 0.28 |
ENST00000253669.5
ENST00000448593.2 |
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr14_+_58894404 | 0.28 |
ENST00000554463.1
ENST00000555833.1 |
KIAA0586
|
KIAA0586 |
| chr20_+_6748311 | 0.28 |
ENST00000378827.4
|
BMP2
|
bone morphogenetic protein 2 |
| chr1_+_212738676 | 0.28 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr21_-_19858196 | 0.28 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr5_+_63461642 | 0.28 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr22_-_30814469 | 0.27 |
ENST00000598426.1
|
KIAA1658
|
KIAA1658 |
| chr9_+_74920335 | 0.27 |
ENST00000451596.2
ENST00000436054.1 |
RP11-63P12.6
|
RP11-63P12.6 |
| chr17_-_42019836 | 0.27 |
ENST00000225992.3
|
PPY
|
pancreatic polypeptide |
| chr18_+_12947126 | 0.27 |
ENST00000592170.1
|
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr8_-_37411648 | 0.27 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr11_-_102668879 | 0.27 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr19_-_38183228 | 0.27 |
ENST00000587199.1
|
ZFP30
|
ZFP30 zinc finger protein |
| chr7_+_125078119 | 0.27 |
ENST00000458437.1
ENST00000415896.1 |
RP11-807H17.1
|
RP11-807H17.1 |
| chr2_+_152214098 | 0.27 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr2_-_191878162 | 0.27 |
ENST00000540176.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr3_-_50340996 | 0.27 |
ENST00000266031.4
ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1
|
hyaluronoglucosaminidase 1 |
| chr22_-_30901637 | 0.27 |
ENST00000381982.3
ENST00000255858.7 ENST00000540456.1 ENST00000392772.2 |
SEC14L4
|
SEC14-like 4 (S. cerevisiae) |
| chr17_-_47786375 | 0.27 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr1_-_94344686 | 0.27 |
ENST00000528680.1
|
DNTTIP2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr19_-_17516449 | 0.27 |
ENST00000252593.6
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr15_-_35838348 | 0.26 |
ENST00000561411.1
ENST00000256538.4 ENST00000440392.2 |
DPH6
|
diphthamine biosynthesis 6 |
| chr7_-_139763521 | 0.26 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr6_-_133035185 | 0.26 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr2_+_228736335 | 0.26 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr19_-_56879416 | 0.26 |
ENST00000591036.1
|
AC006116.21
|
AC006116.21 |
| chr1_-_100643765 | 0.26 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr3_+_109128961 | 0.26 |
ENST00000489670.1
|
RP11-702L6.4
|
RP11-702L6.4 |
| chr6_+_32146268 | 0.26 |
ENST00000427134.2
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr9_+_75766652 | 0.26 |
ENST00000257497.6
|
ANXA1
|
annexin A1 |
| chr18_-_43678241 | 0.26 |
ENST00000593152.2
ENST00000589252.1 ENST00000590665.1 ENST00000398752.6 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr5_-_43397184 | 0.26 |
ENST00000513525.1
|
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr19_-_44809121 | 0.26 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chr19_-_5838768 | 0.26 |
ENST00000527106.1
ENST00000531199.1 ENST00000529165.1 |
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr16_-_58004992 | 0.26 |
ENST00000564448.1
ENST00000251102.8 ENST00000311183.4 |
CNGB1
|
cyclic nucleotide gated channel beta 1 |
| chr2_+_204103733 | 0.25 |
ENST00000443941.1
|
CYP20A1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr19_+_58694396 | 0.25 |
ENST00000326804.4
ENST00000345813.3 ENST00000424679.2 |
ZNF274
|
zinc finger protein 274 |
| chrX_-_106449656 | 0.25 |
ENST00000372466.4
ENST00000421752.1 ENST00000372461.3 |
NUP62CL
|
nucleoporin 62kDa C-terminal like |
| chr12_-_110486281 | 0.25 |
ENST00000546627.1
|
C12orf76
|
chromosome 12 open reading frame 76 |
| chr12_-_49582593 | 0.25 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr15_-_72978490 | 0.25 |
ENST00000311755.3
|
HIGD2B
|
HIG1 hypoxia inducible domain family, member 2B |
| chr4_+_89299885 | 0.25 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr19_+_37808831 | 0.24 |
ENST00000589801.1
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr12_-_52845910 | 0.24 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chr3_-_170626376 | 0.24 |
ENST00000487522.1
ENST00000474417.1 |
EIF5A2
|
eukaryotic translation initiation factor 5A2 |
| chrX_-_153141783 | 0.24 |
ENST00000458029.1
|
L1CAM
|
L1 cell adhesion molecule |
| chr4_+_39640787 | 0.24 |
ENST00000532680.1
|
RP11-539G18.2
|
RP11-539G18.2 |
| chr1_+_155278539 | 0.24 |
ENST00000447866.1
|
FDPS
|
farnesyl diphosphate synthase |
| chr6_+_10694900 | 0.24 |
ENST00000379568.3
|
PAK1IP1
|
PAK1 interacting protein 1 |
| chr4_+_146560245 | 0.24 |
ENST00000541599.1
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr12_+_8850277 | 0.24 |
ENST00000539923.1
ENST00000537189.1 |
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr10_-_1034237 | 0.24 |
ENST00000381466.1
|
AL359878.1
|
Uncharacterized protein |
| chr14_-_58894332 | 0.24 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr18_+_22040593 | 0.24 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr14_+_22591276 | 0.24 |
ENST00000390455.3
|
TRAV26-1
|
T cell receptor alpha variable 26-1 |
| chr10_+_78078088 | 0.24 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chr2_-_240230890 | 0.24 |
ENST00000446876.1
|
HDAC4
|
histone deacetylase 4 |
| chr7_+_90338547 | 0.23 |
ENST00000446790.1
|
CDK14
|
cyclin-dependent kinase 14 |
| chr10_-_49482907 | 0.23 |
ENST00000374201.3
ENST00000407470.4 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr6_-_32908765 | 0.23 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr17_+_4643300 | 0.23 |
ENST00000433935.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr1_-_163172625 | 0.23 |
ENST00000527988.1
ENST00000531476.1 ENST00000530507.1 |
RGS5
|
regulator of G-protein signaling 5 |
| chr16_-_11922665 | 0.23 |
ENST00000573319.1
ENST00000577041.1 ENST00000574028.1 ENST00000571259.1 ENST00000573037.1 ENST00000571158.1 |
BCAR4
|
breast cancer anti-estrogen resistance 4 (non-protein coding) |
| chr19_-_37341673 | 0.23 |
ENST00000586323.1
|
ZNF790
|
zinc finger protein 790 |
| chr12_-_49523896 | 0.23 |
ENST00000549870.1
|
TUBA1B
|
tubulin, alpha 1b |
| chr3_+_42885450 | 0.23 |
ENST00000492609.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr16_+_82068830 | 0.23 |
ENST00000199936.4
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr16_+_67360856 | 0.23 |
ENST00000568804.2
|
LRRC36
|
leucine rich repeat containing 36 |
| chr8_-_131399110 | 0.23 |
ENST00000521426.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr8_+_94767109 | 0.23 |
ENST00000409623.3
ENST00000453906.1 ENST00000521517.1 |
TMEM67
|
transmembrane protein 67 |
| chr5_-_77072085 | 0.23 |
ENST00000518338.2
ENST00000520039.1 ENST00000306388.6 ENST00000520361.1 |
TBCA
|
tubulin folding cofactor A |
| chr17_-_4689727 | 0.23 |
ENST00000328739.5
ENST00000354194.4 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr12_-_106477805 | 0.23 |
ENST00000553094.1
ENST00000549704.1 |
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr6_-_32098013 | 0.22 |
ENST00000375156.3
|
FKBPL
|
FK506 binding protein like |
| chr12_-_86650077 | 0.22 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr3_-_16524357 | 0.22 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr22_-_38480100 | 0.22 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr8_+_99076509 | 0.22 |
ENST00000318528.3
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr4_-_147443043 | 0.22 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr3_-_172312460 | 0.22 |
ENST00000418839.2
|
RP11-408H1.3
|
RP11-408H1.3 |
| chr4_-_170897045 | 0.22 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr12_+_8849773 | 0.22 |
ENST00000541044.1
|
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr18_+_32290218 | 0.22 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr2_-_220119280 | 0.22 |
ENST00000392088.2
|
TUBA4A
|
tubulin, alpha 4a |
| chr18_+_61557781 | 0.22 |
ENST00000443281.1
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr6_+_88054530 | 0.22 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr7_+_137761167 | 0.22 |
ENST00000432161.1
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr19_+_38880252 | 0.22 |
ENST00000586301.1
|
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr3_-_23958402 | 0.22 |
ENST00000415901.2
ENST00000416026.2 ENST00000412028.1 ENST00000388759.3 ENST00000437230.1 |
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1 |
| chr1_+_151009054 | 0.22 |
ENST00000295294.7
|
BNIPL
|
BCL2/adenovirus E1B 19kD interacting protein like |
| chr3_-_170626418 | 0.22 |
ENST00000474096.1
ENST00000295822.2 |
EIF5A2
|
eukaryotic translation initiation factor 5A2 |
| chr22_+_45714301 | 0.22 |
ENST00000427777.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr3_-_160823158 | 0.22 |
ENST00000392779.2
ENST00000392780.1 ENST00000494173.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr5_-_16936340 | 0.21 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr17_+_74536115 | 0.21 |
ENST00000592014.1
|
PRCD
|
progressive rod-cone degeneration |
| chr16_+_22516172 | 0.21 |
ENST00000543407.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr1_+_152956549 | 0.21 |
ENST00000307122.2
|
SPRR1A
|
small proline-rich protein 1A |
| chr19_-_44174330 | 0.21 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr4_-_89079817 | 0.21 |
ENST00000505480.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr5_-_142023766 | 0.21 |
ENST00000394496.2
|
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr20_+_48909240 | 0.21 |
ENST00000371639.3
|
RP11-290F20.1
|
RP11-290F20.1 |
| chr12_+_46777450 | 0.21 |
ENST00000551503.1
|
RP11-96H19.1
|
RP11-96H19.1 |
| chr17_-_63822563 | 0.21 |
ENST00000317442.8
|
CEP112
|
centrosomal protein 112kDa |
| chr12_-_107487604 | 0.21 |
ENST00000008527.5
|
CRY1
|
cryptochrome 1 (photolyase-like) |
| chr16_+_72042487 | 0.21 |
ENST00000572887.1
ENST00000219240.4 ENST00000574309.1 ENST00000576145.1 |
DHODH
|
dihydroorotate dehydrogenase (quinone) |
| chr17_+_33895090 | 0.21 |
ENST00000592381.1
|
RP11-1094M14.11
|
RP11-1094M14.11 |
| chr8_-_102275131 | 0.21 |
ENST00000523121.1
|
KB-1410C5.2
|
KB-1410C5.2 |
| chr11_-_61849656 | 0.21 |
ENST00000524942.1
|
RP11-810P12.5
|
RP11-810P12.5 |
| chr1_-_150849174 | 0.21 |
ENST00000515192.1
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr20_+_25388293 | 0.21 |
ENST00000262460.4
ENST00000429262.2 |
GINS1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr11_+_44587206 | 0.21 |
ENST00000525210.1
ENST00000527737.1 ENST00000524704.1 |
CD82
|
CD82 molecule |
Network of associatons between targets according to the STRING database.
First level regulatory network of PITX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.2 | 0.8 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.2 | 0.6 | GO:0031394 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 0.7 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.2 | 0.5 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.2 | 0.6 | GO:2000909 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of cholesterol import(GO:0060620) regulation of intestinal lipid absorption(GO:1904729) regulation of sterol import(GO:2000909) |
| 0.1 | 0.3 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.3 | GO:0060129 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) regulation of calcium-independent cell-cell adhesion(GO:0051040) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.1 | 0.3 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.4 | GO:2000360 | negative regulation of fertilization(GO:0060467) regulation of binding of sperm to zona pellucida(GO:2000359) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.3 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) |
| 0.1 | 0.4 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.4 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.4 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.6 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 0.6 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.2 | GO:0008078 | mesoderm migration involved in gastrulation(GO:0007509) mesodermal cell migration(GO:0008078) |
| 0.1 | 0.8 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.1 | 0.1 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.1 | 0.9 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.1 | 0.3 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.3 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 0.1 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 0.1 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.1 | 0.2 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.3 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.3 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.2 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.1 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.4 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.3 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.1 | 0.2 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.2 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.7 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.4 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 0.1 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.4 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.4 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.2 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.1 | 0.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 0.5 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.2 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.1 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.6 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 3.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.3 | GO:1903281 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.0 | 0.2 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:2000523 | negative regulation of regulatory T cell differentiation(GO:0045590) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.4 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 1.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.2 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0072221 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.0 | 0.1 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.2 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.0 | GO:2000697 | negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:1904875 | regulation of DNA ligase activity(GO:1904875) |
| 0.0 | 0.4 | GO:0071988 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.2 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.2 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.6 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0042441 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.4 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.2 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.5 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.3 | GO:2000855 | mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.0 | 1.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.2 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.0 | GO:0031000 | response to caffeine(GO:0031000) cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 0.1 | GO:0072387 | flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.9 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:2000302 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.3 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 1.3 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.0 | 0.1 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.0 | 0.2 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.3 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.3 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.2 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.3 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.2 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:2000846 | corticosteroid hormone secretion(GO:0035930) regulation of corticosteroid hormone secretion(GO:2000846) |
| 0.0 | 0.4 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.0 | GO:0033561 | regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.0 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.1 | GO:0007403 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.3 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.0 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.0 | 0.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 0.1 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.4 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.0 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.2 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.0 | GO:0090045 | positive regulation of deacetylase activity(GO:0090045) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.1 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.2 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0060374 | positive regulation of neutrophil apoptotic process(GO:0033031) natural killer cell chemotaxis(GO:0035747) B cell chemotaxis(GO:0035754) mast cell differentiation(GO:0060374) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.3 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.1 | GO:0055013 | cardiac cell development(GO:0055006) cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.2 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.2 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) positive regulation of necroptotic process(GO:0060545) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.6 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.2 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.2 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.2 | GO:0035766 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.2 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.0 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.0 | 0.3 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.0 | 0.2 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.0 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.3 | GO:0051194 | positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.0 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.2 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.2 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.1 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.3 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.2 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.0 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.0 | GO:0048145 | fibroblast proliferation(GO:0048144) regulation of fibroblast proliferation(GO:0048145) |
| 0.0 | 0.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:1903979 | negative regulation of microglial cell activation(GO:1903979) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.0 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.0 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.1 | GO:0033603 | postganglionic parasympathetic fiber development(GO:0021784) positive regulation of dopamine secretion(GO:0033603) positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 0.0 | 0.0 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.4 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.0 | GO:0071810 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.2 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.1 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.0 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0014048 | regulation of glutamate secretion(GO:0014048) |
| 0.0 | 0.0 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0034397 | telomere localization(GO:0034397) meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 | 0.1 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.0 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.0 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.3 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.0 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.1 | GO:0015870 | acetylcholine transport(GO:0015870) otolith morphogenesis(GO:0032474) acetylcholine secretion(GO:0061526) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.0 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.4 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.0 | GO:0046709 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.0 | 0.1 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.0 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) |
| 0.0 | 0.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.0 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.0 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.2 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.0 | GO:0060694 | regulation of cholesterol transporter activity(GO:0060694) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.3 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.1 | 0.3 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.8 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.4 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.2 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.2 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 3.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.1 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.4 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.4 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.5 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.3 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.0 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.0 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.0 | GO:0097180 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0031261 | GINS complex(GO:0000811) DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.0 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.0 | GO:0070685 | macropinocytic cup(GO:0070685) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 0.5 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 0.6 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.1 | 0.6 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.6 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.3 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.1 | 0.4 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.4 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.6 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.4 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.4 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.1 | 0.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.5 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.2 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 1.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.2 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.2 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 0.3 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.1 | GO:0035643 | L-DOPA receptor activity(GO:0035643) L-DOPA binding(GO:0072544) |
| 0.0 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.0 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.0 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.0 | 0.3 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.2 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.2 | GO:1904493 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.0 | 0.1 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.3 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0051731 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.4 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0019144 | ADP-sugar diphosphatase activity(GO:0019144) |
| 0.0 | 0.4 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.1 | GO:0004325 | ferrochelatase activity(GO:0004325) |
| 0.0 | 0.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.0 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.1 | GO:0047946 | glutamine N-acyltransferase activity(GO:0047946) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.3 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0004644 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 1.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0015464 | acetylcholine receptor activity(GO:0015464) G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.0 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0060229 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.0 | 0.1 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.0 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.0 | 0.0 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 1.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0051998 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 2.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 2.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 2.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.9 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |