Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
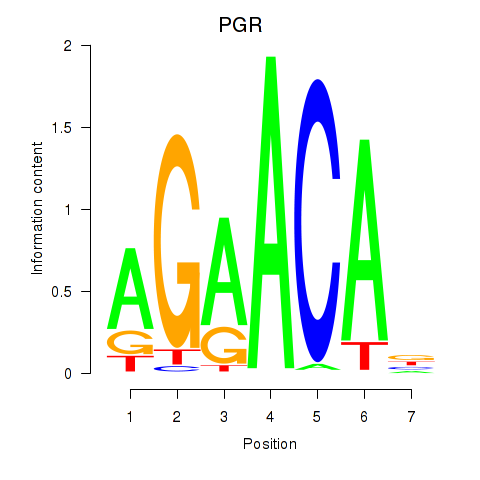
Results for PGR
Z-value: 0.49
Transcription factors associated with PGR
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PGR
|
ENSG00000082175.10 | progesterone receptor |
Activity profile of PGR motif
Sorted Z-values of PGR motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_22766766 | 0.55 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr3_+_52454971 | 0.39 |
ENST00000465863.1
|
PHF7
|
PHD finger protein 7 |
| chr7_-_75241096 | 0.27 |
ENST00000420909.1
|
HIP1
|
huntingtin interacting protein 1 |
| chr17_+_67590125 | 0.24 |
ENST00000591334.1
|
AC003051.1
|
AC003051.1 |
| chr5_+_148443049 | 0.22 |
ENST00000515304.1
ENST00000507318.1 |
CTC-529P8.1
|
CTC-529P8.1 |
| chr17_+_46970134 | 0.22 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr15_+_31658349 | 0.21 |
ENST00000558844.1
|
KLF13
|
Kruppel-like factor 13 |
| chr6_+_30951487 | 0.21 |
ENST00000486149.2
ENST00000376296.3 |
MUC21
|
mucin 21, cell surface associated |
| chr17_-_7760457 | 0.18 |
ENST00000576384.1
|
LSMD1
|
LSM domain containing 1 |
| chr16_+_58497274 | 0.18 |
ENST00000564207.1
|
NDRG4
|
NDRG family member 4 |
| chr12_-_53575120 | 0.17 |
ENST00000542115.1
|
CSAD
|
cysteine sulfinic acid decarboxylase |
| chr17_+_67759813 | 0.17 |
ENST00000587241.1
|
AC003051.1
|
AC003051.1 |
| chr19_-_41903161 | 0.17 |
ENST00000602129.1
ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5
|
exosome component 5 |
| chr17_+_46970127 | 0.16 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr5_-_95018660 | 0.16 |
ENST00000395899.3
ENST00000274432.8 |
SPATA9
|
spermatogenesis associated 9 |
| chr3_-_139195350 | 0.15 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr16_-_28506840 | 0.15 |
ENST00000569430.1
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr17_+_25958174 | 0.15 |
ENST00000313648.6
ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9
|
lectin, galactoside-binding, soluble, 9 |
| chr9_-_34665983 | 0.15 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr7_-_127671674 | 0.15 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr10_+_69865866 | 0.15 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr8_+_40010989 | 0.13 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chrX_-_133792480 | 0.13 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr6_+_27107053 | 0.13 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr17_-_15522826 | 0.13 |
ENST00000395906.3
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr4_+_79567314 | 0.13 |
ENST00000503539.1
ENST00000504675.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr11_-_615570 | 0.13 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr3_-_114035026 | 0.13 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr15_+_67841330 | 0.13 |
ENST00000354498.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chrX_+_109602039 | 0.13 |
ENST00000520821.1
|
RGAG1
|
retrotransposon gag domain containing 1 |
| chr17_-_27188984 | 0.13 |
ENST00000582320.2
|
MIR144
|
microRNA 451b |
| chr1_+_209859510 | 0.12 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr16_+_86600857 | 0.12 |
ENST00000320354.4
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1) |
| chr12_-_122238464 | 0.12 |
ENST00000546227.1
|
RHOF
|
ras homolog family member F (in filopodia) |
| chr20_+_35090150 | 0.12 |
ENST00000340491.4
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4 |
| chr17_+_38119216 | 0.11 |
ENST00000301659.4
|
GSDMA
|
gasdermin A |
| chr17_-_15554940 | 0.11 |
ENST00000455584.2
|
RP11-385D13.1
|
Uncharacterized protein |
| chr15_-_74726283 | 0.11 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr15_-_45406385 | 0.11 |
ENST00000389039.6
|
DUOX2
|
dual oxidase 2 |
| chr12_-_7244469 | 0.11 |
ENST00000538050.1
ENST00000536053.2 |
C1R
|
complement component 1, r subcomponent |
| chr1_-_205290865 | 0.11 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr6_-_75915757 | 0.11 |
ENST00000322507.8
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr2_-_216300784 | 0.11 |
ENST00000421182.1
ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1
|
fibronectin 1 |
| chr13_+_78315466 | 0.10 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr17_+_73997796 | 0.10 |
ENST00000586261.1
|
CDK3
|
cyclin-dependent kinase 3 |
| chr4_+_117220016 | 0.10 |
ENST00000604093.1
|
MTRNR2L13
|
MT-RNR2-like 13 (pseudogene) |
| chr8_-_116681686 | 0.10 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr4_+_79567362 | 0.10 |
ENST00000512322.1
|
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr2_-_220034745 | 0.10 |
ENST00000455516.2
ENST00000396775.3 ENST00000295738.7 |
SLC23A3
|
solute carrier family 23, member 3 |
| chr3_+_125985620 | 0.10 |
ENST00000511512.1
ENST00000512435.1 |
RP11-71E19.1
|
RP11-71E19.1 |
| chr2_+_9778872 | 0.09 |
ENST00000478468.1
|
RP11-521D12.1
|
RP11-521D12.1 |
| chr6_-_154831779 | 0.09 |
ENST00000607772.1
|
CNKSR3
|
CNKSR family member 3 |
| chr14_+_24674926 | 0.09 |
ENST00000339917.5
ENST00000556621.1 ENST00000287913.6 ENST00000428351.2 ENST00000555092.1 |
TSSK4
|
testis-specific serine kinase 4 |
| chr6_-_32908792 | 0.09 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr8_-_143851203 | 0.09 |
ENST00000521396.1
ENST00000317543.7 |
LYNX1
|
Ly6/neurotoxin 1 |
| chr2_-_25451065 | 0.09 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr5_-_121413974 | 0.09 |
ENST00000231004.4
|
LOX
|
lysyl oxidase |
| chr7_-_14026063 | 0.08 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr1_+_41174988 | 0.08 |
ENST00000372652.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr17_+_61086917 | 0.08 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr17_+_46970178 | 0.08 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr15_+_54305101 | 0.08 |
ENST00000260323.11
ENST00000545554.1 ENST00000537900.1 |
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr2_+_64681103 | 0.08 |
ENST00000464281.1
|
LGALSL
|
lectin, galactoside-binding-like |
| chr11_+_123396528 | 0.08 |
ENST00000322282.7
ENST00000529750.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chr6_-_32908765 | 0.08 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr13_+_78315528 | 0.08 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr20_+_25388293 | 0.08 |
ENST00000262460.4
ENST00000429262.2 |
GINS1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr11_-_10920838 | 0.08 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr6_-_24489842 | 0.08 |
ENST00000230036.1
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr5_-_115872142 | 0.08 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr2_+_96991935 | 0.08 |
ENST00000361124.4
ENST00000420728.1 ENST00000542887.1 |
ITPRIPL1
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 1 |
| chr12_-_91505608 | 0.08 |
ENST00000266718.4
|
LUM
|
lumican |
| chr2_-_175711924 | 0.08 |
ENST00000444573.1
|
CHN1
|
chimerin 1 |
| chr2_+_64681219 | 0.08 |
ENST00000238875.5
|
LGALSL
|
lectin, galactoside-binding-like |
| chr9_-_37034028 | 0.08 |
ENST00000520281.1
ENST00000446742.1 ENST00000522003.1 ENST00000523145.1 ENST00000414447.1 ENST00000377847.2 ENST00000377853.2 ENST00000377852.2 ENST00000523241.1 ENST00000520154.1 ENST00000358127.4 |
PAX5
|
paired box 5 |
| chr5_-_180242576 | 0.08 |
ENST00000514438.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr3_+_187871060 | 0.07 |
ENST00000448637.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr8_+_22844913 | 0.07 |
ENST00000519685.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr20_-_25320367 | 0.07 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chr11_-_65625014 | 0.07 |
ENST00000534784.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr6_+_30853002 | 0.07 |
ENST00000421124.2
ENST00000512725.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr2_+_232135245 | 0.07 |
ENST00000446447.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr17_-_46608272 | 0.07 |
ENST00000577092.1
ENST00000239174.6 |
HOXB1
|
homeobox B1 |
| chr4_-_40516560 | 0.07 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chrX_-_130037198 | 0.07 |
ENST00000370935.1
ENST00000338144.3 ENST00000394363.1 |
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr11_-_75379479 | 0.07 |
ENST00000434603.2
|
MAP6
|
microtubule-associated protein 6 |
| chr21_-_28215332 | 0.07 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr11_+_123396307 | 0.07 |
ENST00000456860.2
|
GRAMD1B
|
GRAM domain containing 1B |
| chr21_+_39668831 | 0.07 |
ENST00000419868.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr15_+_31619013 | 0.07 |
ENST00000307145.3
|
KLF13
|
Kruppel-like factor 13 |
| chr7_-_5821314 | 0.07 |
ENST00000425013.2
ENST00000389902.3 |
RNF216
|
ring finger protein 216 |
| chr8_-_26371608 | 0.07 |
ENST00000522362.2
|
PNMA2
|
paraneoplastic Ma antigen 2 |
| chr17_+_41150793 | 0.07 |
ENST00000586277.1
|
RPL27
|
ribosomal protein L27 |
| chr17_+_72667239 | 0.07 |
ENST00000402449.4
|
RAB37
|
RAB37, member RAS oncogene family |
| chr22_+_30279144 | 0.07 |
ENST00000401950.2
ENST00000333027.3 ENST00000445401.1 ENST00000323630.5 ENST00000351488.3 |
MTMR3
|
myotubularin related protein 3 |
| chr5_+_66124590 | 0.07 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_113885138 | 0.07 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr19_-_51529849 | 0.07 |
ENST00000600362.1
ENST00000453757.3 ENST00000601671.1 |
KLK11
|
kallikrein-related peptidase 11 |
| chr11_+_64851666 | 0.07 |
ENST00000525509.1
ENST00000294258.3 ENST00000526334.1 |
ZFPL1
|
zinc finger protein-like 1 |
| chr15_+_94899183 | 0.06 |
ENST00000557742.1
|
MCTP2
|
multiple C2 domains, transmembrane 2 |
| chr1_+_95975672 | 0.06 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr7_-_92855762 | 0.06 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr3_+_185046676 | 0.06 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr5_-_138718973 | 0.06 |
ENST00000353963.3
ENST00000348729.3 |
SLC23A1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr10_+_95372289 | 0.06 |
ENST00000371447.3
|
PDE6C
|
phosphodiesterase 6C, cGMP-specific, cone, alpha prime |
| chr3_-_128294929 | 0.06 |
ENST00000356020.2
|
C3orf27
|
chromosome 3 open reading frame 27 |
| chr6_-_132834184 | 0.06 |
ENST00000367941.2
ENST00000367937.4 |
STX7
|
syntaxin 7 |
| chr13_+_78315348 | 0.06 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr20_-_60573188 | 0.06 |
ENST00000474089.1
|
TAF4
|
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa |
| chrX_-_130037162 | 0.06 |
ENST00000432489.1
|
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr2_+_27440229 | 0.06 |
ENST00000264705.4
ENST00000403525.1 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr2_-_85641162 | 0.06 |
ENST00000447219.2
ENST00000409670.1 ENST00000409724.1 |
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr11_+_64851729 | 0.06 |
ENST00000526791.1
ENST00000526945.1 |
ZFPL1
|
zinc finger protein-like 1 |
| chr16_-_3306587 | 0.06 |
ENST00000541159.1
ENST00000536379.1 ENST00000219596.1 ENST00000339854.4 |
MEFV
|
Mediterranean fever |
| chr2_-_219537134 | 0.06 |
ENST00000295704.2
|
RNF25
|
ring finger protein 25 |
| chr18_+_21529811 | 0.06 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr6_+_143447322 | 0.06 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chr19_+_49377575 | 0.06 |
ENST00000600406.1
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A |
| chr5_-_179107975 | 0.06 |
ENST00000376974.4
|
CBY3
|
chibby homolog 3 (Drosophila) |
| chr4_-_7069760 | 0.06 |
ENST00000264954.4
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli) |
| chr19_-_6333614 | 0.06 |
ENST00000301452.4
|
ACER1
|
alkaline ceramidase 1 |
| chr5_+_139505520 | 0.06 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr8_+_118532937 | 0.06 |
ENST00000297347.3
|
MED30
|
mediator complex subunit 30 |
| chr8_+_70476088 | 0.06 |
ENST00000525999.1
|
SULF1
|
sulfatase 1 |
| chr2_-_163695128 | 0.06 |
ENST00000332142.5
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chrX_+_54834004 | 0.06 |
ENST00000375068.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr6_-_43276535 | 0.05 |
ENST00000372569.3
ENST00000274990.4 |
CRIP3
|
cysteine-rich protein 3 |
| chr4_-_23735183 | 0.05 |
ENST00000507666.1
|
RP11-380P13.2
|
RP11-380P13.2 |
| chr6_+_30852738 | 0.05 |
ENST00000508312.1
ENST00000512336.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr12_+_56473628 | 0.05 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr9_+_82188077 | 0.05 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr1_-_154943212 | 0.05 |
ENST00000368445.5
ENST00000448116.2 ENST00000368449.4 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr6_+_112375462 | 0.05 |
ENST00000361714.1
|
WISP3
|
WNT1 inducible signaling pathway protein 3 |
| chr17_+_22022437 | 0.05 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr11_-_65625330 | 0.05 |
ENST00000531407.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr4_-_105416039 | 0.05 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr11_-_6502580 | 0.05 |
ENST00000423813.2
ENST00000396777.3 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr1_+_152691998 | 0.05 |
ENST00000368775.2
|
C1orf68
|
chromosome 1 open reading frame 68 |
| chr4_-_87770416 | 0.05 |
ENST00000273905.6
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter), member 6 |
| chr7_-_27213893 | 0.05 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr11_-_6502534 | 0.05 |
ENST00000254584.2
ENST00000525235.1 ENST00000445086.2 |
ARFIP2
|
ADP-ribosylation factor interacting protein 2 |
| chr11_-_62358972 | 0.05 |
ENST00000278279.3
|
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chrX_+_46940254 | 0.05 |
ENST00000336169.3
|
RGN
|
regucalcin |
| chr10_+_97709725 | 0.05 |
ENST00000472454.2
|
RP11-248J23.6
|
Protein LOC100652732 |
| chrX_-_100641155 | 0.05 |
ENST00000372880.1
ENST00000308731.7 |
BTK
|
Bruton agammaglobulinemia tyrosine kinase |
| chr5_+_138629628 | 0.05 |
ENST00000508689.1
ENST00000514528.1 |
MATR3
|
matrin 3 |
| chr12_+_27398584 | 0.05 |
ENST00000543246.1
|
STK38L
|
serine/threonine kinase 38 like |
| chr14_-_75330537 | 0.05 |
ENST00000556084.2
ENST00000556489.2 ENST00000445876.1 |
PROX2
|
prospero homeobox 2 |
| chr4_-_88450612 | 0.05 |
ENST00000418378.1
ENST00000282470.6 |
SPARCL1
|
SPARC-like 1 (hevin) |
| chr2_+_105050794 | 0.05 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr5_-_137374288 | 0.05 |
ENST00000514310.1
|
FAM13B
|
family with sequence similarity 13, member B |
| chr12_-_71551652 | 0.05 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr11_+_65627974 | 0.05 |
ENST00000525768.1
|
MUS81
|
MUS81 structure-specific endonuclease subunit |
| chr2_-_175711978 | 0.05 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chrX_+_69672136 | 0.05 |
ENST00000374355.3
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr3_-_114343039 | 0.05 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chrX_+_23352133 | 0.05 |
ENST00000379361.4
|
PTCHD1
|
patched domain containing 1 |
| chrX_+_47093171 | 0.05 |
ENST00000377078.2
|
USP11
|
ubiquitin specific peptidase 11 |
| chr11_-_26744908 | 0.05 |
ENST00000533617.1
|
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr20_+_23420885 | 0.05 |
ENST00000246020.2
|
CSTL1
|
cystatin-like 1 |
| chr11_-_88796803 | 0.05 |
ENST00000418177.2
ENST00000455756.2 |
GRM5
|
glutamate receptor, metabotropic 5 |
| chr6_-_159466136 | 0.05 |
ENST00000367066.3
ENST00000326965.6 |
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr20_-_52612468 | 0.05 |
ENST00000422805.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr7_-_129845313 | 0.05 |
ENST00000397622.2
|
TMEM209
|
transmembrane protein 209 |
| chr6_-_128222103 | 0.04 |
ENST00000434358.1
ENST00000543064.1 ENST00000368248.2 |
THEMIS
|
thymocyte selection associated |
| chr8_+_124968225 | 0.04 |
ENST00000399018.1
|
FER1L6
|
fer-1-like 6 (C. elegans) |
| chr1_+_41447609 | 0.04 |
ENST00000543104.1
|
CTPS1
|
CTP synthase 1 |
| chr9_-_34729457 | 0.04 |
ENST00000378788.3
|
FAM205A
|
family with sequence similarity 205, member A |
| chr10_-_104178857 | 0.04 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr14_+_23709555 | 0.04 |
ENST00000430154.2
|
C14orf164
|
chromosome 14 open reading frame 164 |
| chr20_+_31446729 | 0.04 |
ENST00000400522.4
|
EFCAB8
|
EF-hand calcium binding domain 8 |
| chr3_-_114343768 | 0.04 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr6_+_167412835 | 0.04 |
ENST00000349556.4
|
FGFR1OP
|
FGFR1 oncogene partner |
| chr10_+_90562705 | 0.04 |
ENST00000539337.1
|
LIPM
|
lipase, family member M |
| chr1_-_154943002 | 0.04 |
ENST00000606391.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr11_-_115127611 | 0.04 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr19_+_19431490 | 0.04 |
ENST00000392313.6
ENST00000262815.8 ENST00000609122.1 |
MAU2
|
MAU2 sister chromatid cohesion factor |
| chr5_+_138629337 | 0.04 |
ENST00000394805.3
ENST00000512876.1 ENST00000513678.1 |
MATR3
|
matrin 3 |
| chr17_-_7760779 | 0.04 |
ENST00000335155.5
ENST00000575071.1 |
LSMD1
|
LSM domain containing 1 |
| chr19_-_48059113 | 0.04 |
ENST00000391901.3
ENST00000314121.4 ENST00000448976.1 |
ZNF541
|
zinc finger protein 541 |
| chr7_-_37026108 | 0.04 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr19_-_19144243 | 0.04 |
ENST00000594445.1
ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2
|
SURP and G patch domain containing 2 |
| chr8_-_67974552 | 0.04 |
ENST00000357849.4
|
COPS5
|
COP9 signalosome subunit 5 |
| chr6_+_108487245 | 0.04 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chrX_-_78622805 | 0.04 |
ENST00000373298.2
|
ITM2A
|
integral membrane protein 2A |
| chr16_+_50582222 | 0.04 |
ENST00000268459.3
|
NKD1
|
naked cuticle homolog 1 (Drosophila) |
| chr12_+_53848505 | 0.04 |
ENST00000552819.1
ENST00000455667.3 |
PCBP2
|
poly(rC) binding protein 2 |
| chr2_-_158182410 | 0.04 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr10_+_90562487 | 0.04 |
ENST00000404743.4
|
LIPM
|
lipase, family member M |
| chr6_-_10413112 | 0.04 |
ENST00000465858.1
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr20_+_34129770 | 0.04 |
ENST00000348547.2
ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3
|
ERGIC and golgi 3 |
| chr17_+_7761013 | 0.04 |
ENST00000571846.1
|
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr5_-_156536126 | 0.04 |
ENST00000522593.1
|
HAVCR2
|
hepatitis A virus cellular receptor 2 |
| chr3_+_169539710 | 0.04 |
ENST00000340806.6
|
LRRIQ4
|
leucine-rich repeats and IQ motif containing 4 |
| chr4_+_79567057 | 0.04 |
ENST00000503259.1
ENST00000507802.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr3_+_137906109 | 0.04 |
ENST00000481646.1
ENST00000469044.1 ENST00000491704.1 ENST00000461600.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr6_-_53013620 | 0.04 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr4_-_71532207 | 0.04 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr17_+_65027509 | 0.04 |
ENST00000375684.1
|
AC005544.1
|
Uncharacterized protein |
| chr7_+_73442457 | 0.04 |
ENST00000438880.1
ENST00000414324.1 ENST00000380562.4 |
ELN
|
elastin |
Network of associatons between targets according to the STRING database.
First level regulatory network of PGR
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.1 | 0.3 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.3 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.2 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.1 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) glomerular endothelium development(GO:0072011) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.1 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.1 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.0 | GO:0002519 | natural killer cell tolerance induction(GO:0002519) |
| 0.0 | 0.1 | GO:0010982 | negative regulation of triglyceride catabolic process(GO:0010897) regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0051510 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:1903625 | negative regulation of sperm motility(GO:1901318) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.1 | GO:0002368 | B cell cytokine production(GO:0002368) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.0 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.0 | GO:0002118 | aggressive behavior(GO:0002118) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.1 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.1 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.0 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST STAT3 PATHWAY | STAT3 Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |