Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
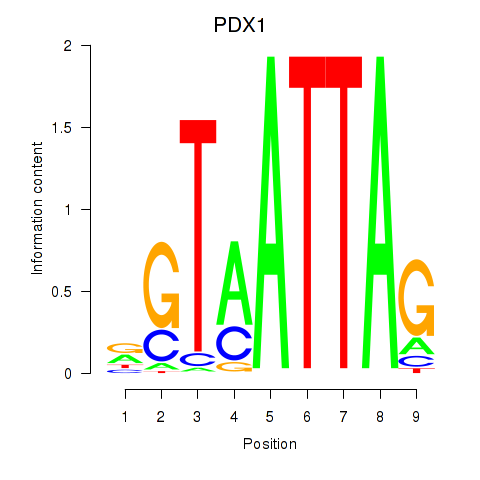
Results for PDX1
Z-value: 0.64
Transcription factors associated with PDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PDX1
|
ENSG00000139515.5 | pancreatic and duodenal homeobox 1 |
Activity profile of PDX1 motif
Sorted Z-values of PDX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_37723336 | 0.62 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr12_-_86650077 | 0.62 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr5_+_66300464 | 0.41 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_+_43968306 | 0.40 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr5_-_56778635 | 0.35 |
ENST00000423391.1
|
ACTBL2
|
actin, beta-like 2 |
| chr2_+_66918558 | 0.35 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr17_-_77924627 | 0.30 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr5_+_66300446 | 0.29 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr8_+_32579321 | 0.28 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr9_-_5339873 | 0.27 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr1_-_152386732 | 0.26 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr15_+_63188009 | 0.25 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr8_+_32579341 | 0.25 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr4_+_71384257 | 0.23 |
ENST00000339336.4
|
AMTN
|
amelotin |
| chr3_+_111718173 | 0.21 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr7_+_129015484 | 0.21 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr12_-_52828147 | 0.21 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chr13_-_30683005 | 0.20 |
ENST00000413591.1
ENST00000432770.1 |
LINC00365
|
long intergenic non-protein coding RNA 365 |
| chr2_+_45168875 | 0.20 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr11_+_110001723 | 0.20 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr2_+_138721850 | 0.20 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr14_-_69261310 | 0.20 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr13_+_78315466 | 0.19 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr4_+_71384300 | 0.18 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr5_+_172068232 | 0.18 |
ENST00000520919.1
ENST00000522853.1 ENST00000369800.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr1_-_149859466 | 0.18 |
ENST00000331128.3
|
HIST2H2AB
|
histone cluster 2, H2ab |
| chr12_-_54813229 | 0.17 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr20_-_31124186 | 0.17 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr8_-_131399110 | 0.17 |
ENST00000521426.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr18_-_46784778 | 0.17 |
ENST00000582399.1
|
DYM
|
dymeclin |
| chr7_-_92777606 | 0.16 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr17_-_38928414 | 0.16 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr8_+_32579271 | 0.16 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr12_-_56321397 | 0.16 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chrX_+_10126488 | 0.15 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr13_+_78315528 | 0.15 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr4_+_147145709 | 0.15 |
ENST00000504313.1
|
RP11-6L6.2
|
Uncharacterized protein |
| chr20_+_43990576 | 0.15 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr12_-_94673956 | 0.15 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr3_-_98235962 | 0.14 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr3_-_141747950 | 0.14 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr3_-_100565249 | 0.14 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr1_-_169396646 | 0.14 |
ENST00000367806.3
|
CCDC181
|
coiled-coil domain containing 181 |
| chr14_+_59100774 | 0.14 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr5_-_115890554 | 0.14 |
ENST00000509665.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr10_-_135379132 | 0.14 |
ENST00000343131.5
|
SYCE1
|
synaptonemal complex central element protein 1 |
| chr15_+_67841330 | 0.13 |
ENST00000354498.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr14_+_72052983 | 0.13 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr15_+_75080883 | 0.13 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr7_+_129015671 | 0.13 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr12_+_49621658 | 0.13 |
ENST00000541364.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr6_-_167040693 | 0.13 |
ENST00000366863.2
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr2_+_228736335 | 0.13 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr9_-_5304432 | 0.13 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr4_-_109541539 | 0.13 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr6_+_126221034 | 0.12 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr20_-_50419055 | 0.12 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr12_-_16761007 | 0.12 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_+_199996702 | 0.12 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr3_+_111717511 | 0.12 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr15_+_59279851 | 0.12 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr3_+_111718036 | 0.12 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr12_+_47610315 | 0.12 |
ENST00000548348.1
ENST00000549500.1 |
PCED1B
|
PC-esterase domain containing 1B |
| chr1_+_107683644 | 0.11 |
ENST00000370067.1
|
NTNG1
|
netrin G1 |
| chr3_+_111717600 | 0.11 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr17_-_40337470 | 0.11 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr7_-_111424462 | 0.11 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr4_+_77356248 | 0.11 |
ENST00000296043.6
|
SHROOM3
|
shroom family member 3 |
| chr6_+_13272904 | 0.11 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr17_+_74846230 | 0.11 |
ENST00000592919.1
|
LINC00868
|
long intergenic non-protein coding RNA 868 |
| chr4_+_53525573 | 0.11 |
ENST00000503051.1
|
USP46-AS1
|
USP46 antisense RNA 1 |
| chr7_-_100860851 | 0.11 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr19_-_7698599 | 0.11 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr2_-_166930131 | 0.11 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr3_+_108541545 | 0.10 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr4_-_103749179 | 0.10 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr15_-_100882191 | 0.10 |
ENST00000268070.4
|
ADAMTS17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chrX_-_106146547 | 0.10 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr18_+_3447572 | 0.10 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chrX_-_19817869 | 0.10 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr17_-_8301132 | 0.10 |
ENST00000399398.2
|
RNF222
|
ring finger protein 222 |
| chr17_-_39165366 | 0.10 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr11_+_65554493 | 0.10 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr11_+_15136462 | 0.10 |
ENST00000379556.3
ENST00000424273.1 |
INSC
|
inscuteable homolog (Drosophila) |
| chr19_+_11071546 | 0.10 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr11_+_115498761 | 0.09 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr11_-_36619771 | 0.09 |
ENST00000311485.3
ENST00000527033.1 ENST00000532616.1 |
RAG2
|
recombination activating gene 2 |
| chr12_-_51611477 | 0.09 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr11_-_129062093 | 0.09 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr21_+_17442799 | 0.09 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_28586006 | 0.09 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chr21_+_17792672 | 0.09 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr7_-_99716940 | 0.09 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr11_-_115630900 | 0.09 |
ENST00000537070.1
ENST00000499809.1 ENST00000514294.2 ENST00000535683.1 |
LINC00900
|
long intergenic non-protein coding RNA 900 |
| chr13_-_46626820 | 0.08 |
ENST00000428921.1
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chrX_-_110655306 | 0.08 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr1_+_28261533 | 0.08 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr17_+_72931876 | 0.08 |
ENST00000328801.4
|
OTOP3
|
otopetrin 3 |
| chr4_+_22999152 | 0.08 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr1_-_221509638 | 0.08 |
ENST00000439004.1
|
RP11-421L10.1
|
RP11-421L10.1 |
| chr2_+_48541776 | 0.08 |
ENST00000413569.1
ENST00000340553.3 |
FOXN2
|
forkhead box N2 |
| chr9_+_131037623 | 0.08 |
ENST00000495313.1
ENST00000372898.2 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr6_-_100912785 | 0.08 |
ENST00000369208.3
|
SIM1
|
single-minded family bHLH transcription factor 1 |
| chr11_+_125034586 | 0.08 |
ENST00000298282.9
|
PKNOX2
|
PBX/knotted 1 homeobox 2 |
| chr8_-_42234745 | 0.08 |
ENST00000220812.2
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr4_-_70518941 | 0.08 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr12_-_6233828 | 0.08 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr12_-_16761117 | 0.08 |
ENST00000538051.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr8_+_22844913 | 0.08 |
ENST00000519685.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr15_+_28624878 | 0.08 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr4_-_138453606 | 0.07 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr9_-_28670283 | 0.07 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr20_+_60174827 | 0.07 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr1_-_2458026 | 0.07 |
ENST00000435556.3
ENST00000378466.3 |
PANK4
|
pantothenate kinase 4 |
| chr6_-_25042231 | 0.07 |
ENST00000510784.2
|
FAM65B
|
family with sequence similarity 65, member B |
| chr11_-_31531121 | 0.07 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr1_-_21625486 | 0.07 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chrX_+_133930798 | 0.07 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr7_+_99717230 | 0.07 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr18_+_55888767 | 0.07 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr1_+_36621174 | 0.07 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr1_+_36621529 | 0.07 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr8_+_77593448 | 0.07 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr20_-_50418947 | 0.07 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr3_-_131756559 | 0.06 |
ENST00000505957.1
|
CPNE4
|
copine IV |
| chr1_+_151735431 | 0.06 |
ENST00000321531.5
ENST00000315067.8 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr20_+_62795827 | 0.06 |
ENST00000328439.1
ENST00000536311.1 |
MYT1
|
myelin transcription factor 1 |
| chr19_-_7968427 | 0.06 |
ENST00000539278.1
|
AC010336.1
|
Uncharacterized protein |
| chr2_-_208031943 | 0.06 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr8_+_55528627 | 0.06 |
ENST00000220676.1
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr4_-_119759795 | 0.06 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr10_+_24497704 | 0.06 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr5_-_54988559 | 0.06 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr15_+_43886057 | 0.06 |
ENST00000441322.1
ENST00000413657.2 ENST00000453733.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr1_+_160160283 | 0.06 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr5_-_140998616 | 0.06 |
ENST00000389054.3
ENST00000398562.2 ENST00000389057.5 ENST00000398566.3 ENST00000398557.4 ENST00000253811.6 |
DIAPH1
|
diaphanous-related formin 1 |
| chr14_+_100485712 | 0.06 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr12_+_81101277 | 0.06 |
ENST00000228641.3
|
MYF6
|
myogenic factor 6 (herculin) |
| chrX_-_64196351 | 0.06 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr15_+_43986069 | 0.06 |
ENST00000415044.1
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr18_+_59000815 | 0.06 |
ENST00000262717.4
|
CDH20
|
cadherin 20, type 2 |
| chr3_+_69985792 | 0.06 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr11_-_8285405 | 0.05 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr2_-_145277569 | 0.05 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr1_+_1846519 | 0.05 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr7_-_14028488 | 0.05 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr7_-_33102338 | 0.05 |
ENST00000610140.1
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chrX_-_100129128 | 0.05 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr1_-_94147385 | 0.05 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr12_+_72332618 | 0.05 |
ENST00000333850.3
|
TPH2
|
tryptophan hydroxylase 2 |
| chr4_-_41884620 | 0.05 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr7_-_33102399 | 0.05 |
ENST00000242210.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr3_-_112127981 | 0.05 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr7_+_114055052 | 0.05 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr14_-_47351391 | 0.05 |
ENST00000399222.3
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr17_+_43238438 | 0.05 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr20_+_30697298 | 0.05 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr15_-_55562582 | 0.05 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr11_-_8290263 | 0.05 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr1_-_1709845 | 0.05 |
ENST00000341426.5
ENST00000344463.4 |
NADK
|
NAD kinase |
| chr8_-_10512569 | 0.05 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr1_+_160160346 | 0.05 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr9_-_16728161 | 0.05 |
ENST00000603713.1
ENST00000603313.1 |
BNC2
|
basonuclin 2 |
| chr12_-_91576429 | 0.05 |
ENST00000552145.1
ENST00000546745.1 |
DCN
|
decorin |
| chr20_-_50418972 | 0.05 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr13_-_67802549 | 0.05 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr17_-_27418537 | 0.04 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr12_-_21928515 | 0.04 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr5_+_98109322 | 0.04 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr12_+_56325231 | 0.04 |
ENST00000549368.1
|
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chrM_+_9207 | 0.04 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr5_-_140998481 | 0.04 |
ENST00000518047.1
|
DIAPH1
|
diaphanous-related formin 1 |
| chr14_+_74034310 | 0.04 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr5_-_96478466 | 0.04 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr1_-_151431674 | 0.04 |
ENST00000531094.1
|
POGZ
|
pogo transposable element with ZNF domain |
| chr7_+_90012986 | 0.04 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr2_-_227050079 | 0.04 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr4_-_103749205 | 0.04 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr10_+_119302508 | 0.04 |
ENST00000442245.4
|
EMX2
|
empty spiracles homeobox 2 |
| chr15_+_43985725 | 0.04 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr5_+_60933634 | 0.04 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr6_+_34204642 | 0.04 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr19_+_13049413 | 0.04 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
| chr21_-_31538971 | 0.04 |
ENST00000286808.3
|
CLDN17
|
claudin 17 |
| chr21_+_17443434 | 0.04 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_-_201140673 | 0.04 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr14_+_61654271 | 0.04 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr12_-_14849470 | 0.03 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chrX_-_64196307 | 0.03 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr12_+_15699286 | 0.03 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr19_+_641178 | 0.03 |
ENST00000166133.3
|
FGF22
|
fibroblast growth factor 22 |
| chr21_-_35899113 | 0.03 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr4_-_66536057 | 0.03 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr3_-_74570291 | 0.03 |
ENST00000263665.6
|
CNTN3
|
contactin 3 (plasmacytoma associated) |
| chr16_+_24621546 | 0.03 |
ENST00000566108.1
|
CTD-2540M10.1
|
CTD-2540M10.1 |
| chr8_+_38831683 | 0.03 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr15_-_31393910 | 0.03 |
ENST00000397795.2
ENST00000256552.6 ENST00000559179.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr15_-_65407524 | 0.03 |
ENST00000559089.1
|
UBAP1L
|
ubiquitin associated protein 1-like |
| chr10_-_27529486 | 0.03 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr20_+_43538692 | 0.03 |
ENST00000217074.4
ENST00000255136.3 |
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of PDX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.4 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0045726 | regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.0 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.0 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.0 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |