Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
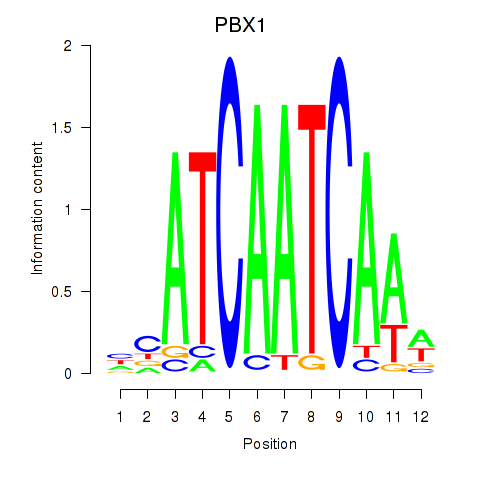
Results for PBX1
Z-value: 0.68
Transcription factors associated with PBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PBX1
|
ENSG00000185630.14 | PBX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PBX1 | hg19_v2_chr1_+_164600184_164600218 | -0.50 | 3.1e-01 | Click! |
Activity profile of PBX1 motif
Sorted Z-values of PBX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_50981079 | 0.68 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34 |
| chr22_-_30642728 | 0.60 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr6_-_26250835 | 0.60 |
ENST00000446824.2
|
HIST1H3F
|
histone cluster 1, H3f |
| chr6_+_27775899 | 0.37 |
ENST00000358739.3
|
HIST1H2AI
|
histone cluster 1, H2ai |
| chr22_-_30642782 | 0.37 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr22_+_29138013 | 0.31 |
ENST00000216027.3
ENST00000398941.2 |
HSCB
|
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr9_+_140135665 | 0.30 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr14_-_24584138 | 0.27 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr2_+_234104079 | 0.25 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr6_+_26199737 | 0.25 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr16_-_88729473 | 0.23 |
ENST00000301012.3
ENST00000569177.1 |
MVD
|
mevalonate (diphospho) decarboxylase |
| chr19_-_5903714 | 0.22 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr11_+_64794991 | 0.22 |
ENST00000352068.5
ENST00000525648.1 |
SNX15
|
sorting nexin 15 |
| chr10_-_18948208 | 0.21 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr14_+_50065459 | 0.21 |
ENST00000318317.4
|
LRR1
|
leucine rich repeat protein 1 |
| chr16_-_20702578 | 0.20 |
ENST00000307493.4
ENST00000219151.4 |
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr19_+_46498704 | 0.20 |
ENST00000595358.1
ENST00000594672.1 ENST00000536603.1 |
CCDC61
|
coiled-coil domain containing 61 |
| chr3_+_193853927 | 0.20 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr10_+_51549498 | 0.20 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr6_-_26032288 | 0.19 |
ENST00000244661.2
|
HIST1H3B
|
histone cluster 1, H3b |
| chr19_-_49121054 | 0.19 |
ENST00000546623.1
ENST00000084795.5 |
RPL18
|
ribosomal protein L18 |
| chr4_-_105416039 | 0.19 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr4_-_69536346 | 0.18 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr12_-_53207842 | 0.18 |
ENST00000458244.2
|
KRT4
|
keratin 4 |
| chr10_+_90484301 | 0.18 |
ENST00000404190.1
|
LIPK
|
lipase, family member K |
| chr6_-_26124138 | 0.18 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr3_+_52570610 | 0.18 |
ENST00000307106.3
ENST00000477703.1 ENST00000476842.1 |
SMIM4
|
small integral membrane protein 4 |
| chr12_-_106477805 | 0.18 |
ENST00000553094.1
ENST00000549704.1 |
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr9_-_32573130 | 0.18 |
ENST00000350021.2
ENST00000379847.3 |
NDUFB6
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa |
| chrX_+_109602039 | 0.18 |
ENST00000520821.1
|
RGAG1
|
retrotransposon gag domain containing 1 |
| chr7_+_120629653 | 0.17 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr16_-_838329 | 0.17 |
ENST00000563560.1
ENST00000569601.1 ENST00000565809.1 ENST00000565377.1 ENST00000007264.2 ENST00000567114.1 |
RPUSD1
|
RNA pseudouridylate synthase domain containing 1 |
| chr5_+_159656437 | 0.17 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chrX_+_125953746 | 0.17 |
ENST00000371125.3
|
CXorf64
|
chromosome X open reading frame 64 |
| chr12_+_81471816 | 0.17 |
ENST00000261206.3
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3 |
| chr7_-_27142290 | 0.17 |
ENST00000222718.5
|
HOXA2
|
homeobox A2 |
| chr5_+_3596168 | 0.16 |
ENST00000302006.3
|
IRX1
|
iroquois homeobox 1 |
| chr19_-_49955050 | 0.15 |
ENST00000262265.5
|
PIH1D1
|
PIH1 domain containing 1 |
| chr19_+_10828724 | 0.15 |
ENST00000585892.1
ENST00000314646.5 ENST00000359692.6 |
DNM2
|
dynamin 2 |
| chr6_-_26199499 | 0.15 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr3_-_112564797 | 0.15 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr11_+_114166536 | 0.14 |
ENST00000299964.3
|
NNMT
|
nicotinamide N-methyltransferase |
| chr2_+_12857015 | 0.14 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr10_+_69865866 | 0.14 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr11_-_2162162 | 0.14 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr10_+_69869237 | 0.13 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr2_+_12857043 | 0.13 |
ENST00000381465.2
|
TRIB2
|
tribbles pseudokinase 2 |
| chr9_-_131486367 | 0.13 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr14_-_64971893 | 0.13 |
ENST00000555220.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr6_-_110736742 | 0.13 |
ENST00000368924.3
ENST00000368923.3 |
DDO
|
D-aspartate oxidase |
| chr15_-_60683326 | 0.13 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr4_+_77356248 | 0.12 |
ENST00000296043.6
|
SHROOM3
|
shroom family member 3 |
| chr5_-_141703713 | 0.12 |
ENST00000511815.1
|
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr7_+_134331550 | 0.12 |
ENST00000344924.3
ENST00000418040.1 ENST00000393132.2 |
BPGM
|
2,3-bisphosphoglycerate mutase |
| chr19_+_19030478 | 0.12 |
ENST00000247003.4
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr19_-_11266437 | 0.12 |
ENST00000586708.1
ENST00000591396.1 ENST00000592967.1 ENST00000585486.1 ENST00000585567.1 |
SPC24
|
SPC24, NDC80 kinetochore complex component |
| chr11_+_844406 | 0.12 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr10_+_124320195 | 0.12 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr16_-_63651939 | 0.12 |
ENST00000563855.1
ENST00000568932.1 ENST00000564290.1 |
RP11-368L12.1
|
RP11-368L12.1 |
| chr19_+_41698927 | 0.12 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr2_+_219110149 | 0.12 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr12_+_48722763 | 0.11 |
ENST00000335017.1
|
H1FNT
|
H1 histone family, member N, testis-specific |
| chr11_-_118966167 | 0.11 |
ENST00000530167.1
|
H2AFX
|
H2A histone family, member X |
| chr17_-_15519008 | 0.11 |
ENST00000261644.8
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr12_-_91573249 | 0.11 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr6_-_36725157 | 0.11 |
ENST00000393189.2
|
CPNE5
|
copine V |
| chrX_+_1734051 | 0.11 |
ENST00000381229.4
ENST00000381233.3 |
ASMT
|
acetylserotonin O-methyltransferase |
| chr19_+_19030497 | 0.11 |
ENST00000438170.2
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr2_+_62132800 | 0.11 |
ENST00000538736.1
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr14_+_23709555 | 0.10 |
ENST00000430154.2
|
C14orf164
|
chromosome 14 open reading frame 164 |
| chr11_+_44117219 | 0.10 |
ENST00000532479.1
ENST00000527014.1 |
EXT2
|
exostosin glycosyltransferase 2 |
| chr8_-_91095099 | 0.10 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr6_-_167040731 | 0.10 |
ENST00000265678.4
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr1_+_205473720 | 0.10 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr3_-_71114066 | 0.10 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
| chr14_+_50065376 | 0.10 |
ENST00000298288.6
|
LRR1
|
leucine rich repeat protein 1 |
| chr2_-_190044480 | 0.10 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chrX_-_1331527 | 0.10 |
ENST00000381567.3
ENST00000381566.1 ENST00000400841.2 |
CRLF2
|
cytokine receptor-like factor 2 |
| chrX_+_140084756 | 0.10 |
ENST00000449283.1
|
SPANXB2
|
SPANX family, member B2 |
| chr9_-_14308004 | 0.10 |
ENST00000493697.1
|
NFIB
|
nuclear factor I/B |
| chr18_-_56985776 | 0.09 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr15_-_44069513 | 0.09 |
ENST00000433927.1
|
ELL3
|
elongation factor RNA polymerase II-like 3 |
| chr18_+_44526786 | 0.09 |
ENST00000245121.5
ENST00000356157.7 |
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr4_+_187148556 | 0.09 |
ENST00000264690.6
ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1 |
| chr1_-_3713042 | 0.09 |
ENST00000378251.1
|
LRRC47
|
leucine rich repeat containing 47 |
| chr15_+_44069276 | 0.09 |
ENST00000381359.1
|
SERF2
|
small EDRK-rich factor 2 |
| chr17_+_8191815 | 0.09 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr7_-_102184083 | 0.09 |
ENST00000379357.5
|
POLR2J3
|
polymerase (RNA) II (DNA directed) polypeptide J3 |
| chr8_-_37411648 | 0.09 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chrX_+_139791917 | 0.09 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr19_+_49999631 | 0.08 |
ENST00000270625.2
ENST00000596873.1 ENST00000594493.1 ENST00000599561.1 |
RPS11
|
ribosomal protein S11 |
| chr7_-_559853 | 0.08 |
ENST00000405692.2
|
PDGFA
|
platelet-derived growth factor alpha polypeptide |
| chr17_+_72983674 | 0.08 |
ENST00000337231.5
|
CDR2L
|
cerebellar degeneration-related protein 2-like |
| chr21_+_39668478 | 0.08 |
ENST00000398927.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr2_+_201997595 | 0.08 |
ENST00000470178.2
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr19_+_12203100 | 0.08 |
ENST00000596883.1
|
ZNF788
|
zinc finger family member 788 |
| chr3_-_161090660 | 0.08 |
ENST00000359175.4
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr7_-_72971934 | 0.08 |
ENST00000411832.1
|
BCL7B
|
B-cell CLL/lymphoma 7B |
| chr20_-_45976816 | 0.07 |
ENST00000441977.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr17_-_40828969 | 0.07 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chrM_+_10053 | 0.07 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr6_-_167040693 | 0.07 |
ENST00000366863.2
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr22_-_22337146 | 0.07 |
ENST00000398793.2
|
TOP3B
|
topoisomerase (DNA) III beta |
| chr8_+_99076750 | 0.07 |
ENST00000545282.1
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr19_-_39881669 | 0.07 |
ENST00000221266.7
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr11_-_32816156 | 0.07 |
ENST00000531481.1
ENST00000335185.5 |
CCDC73
|
coiled-coil domain containing 73 |
| chr19_+_782755 | 0.07 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr22_+_35695793 | 0.07 |
ENST00000456128.1
ENST00000449058.2 ENST00000411850.1 ENST00000425375.1 ENST00000436462.2 ENST00000382034.5 |
TOM1
|
target of myb1 (chicken) |
| chr18_-_14132422 | 0.07 |
ENST00000589498.1
ENST00000590202.1 |
ZNF519
|
zinc finger protein 519 |
| chr11_+_61717279 | 0.07 |
ENST00000378043.4
|
BEST1
|
bestrophin 1 |
| chr6_+_151561085 | 0.07 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr1_-_206288647 | 0.07 |
ENST00000331555.5
|
C1orf186
|
chromosome 1 open reading frame 186 |
| chr19_+_5681153 | 0.07 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr5_+_166711804 | 0.07 |
ENST00000518659.1
ENST00000545108.1 |
TENM2
|
teneurin transmembrane protein 2 |
| chr3_-_149510553 | 0.07 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr22_-_22337204 | 0.06 |
ENST00000430142.1
ENST00000357179.5 |
TOP3B
|
topoisomerase (DNA) III beta |
| chr17_+_78518617 | 0.06 |
ENST00000537330.1
ENST00000570891.1 |
RPTOR
|
regulatory associated protein of MTOR, complex 1 |
| chr11_+_61717336 | 0.06 |
ENST00000378042.3
|
BEST1
|
bestrophin 1 |
| chr5_-_171881362 | 0.06 |
ENST00000519643.1
|
SH3PXD2B
|
SH3 and PX domains 2B |
| chr6_-_27114577 | 0.06 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chr4_-_57844989 | 0.06 |
ENST00000264230.4
|
NOA1
|
nitric oxide associated 1 |
| chr9_-_123676827 | 0.06 |
ENST00000546084.1
|
TRAF1
|
TNF receptor-associated factor 1 |
| chr11_-_126870655 | 0.06 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr4_-_185776789 | 0.06 |
ENST00000510284.1
|
RP11-701P16.5
|
RP11-701P16.5 |
| chr10_+_24738355 | 0.06 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr16_+_19078911 | 0.06 |
ENST00000321998.5
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chrX_+_53123314 | 0.06 |
ENST00000605526.1
ENST00000604062.1 ENST00000604369.1 ENST00000366185.2 ENST00000604849.1 |
RP11-258C19.5
|
long intergenic non-protein coding RNA 1155 |
| chr3_-_178103144 | 0.06 |
ENST00000417383.1
ENST00000418585.1 ENST00000411727.1 ENST00000439810.1 |
RP11-33A14.1
|
RP11-33A14.1 |
| chr11_-_34938039 | 0.06 |
ENST00000395787.3
|
APIP
|
APAF1 interacting protein |
| chr5_+_98109322 | 0.06 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr1_-_54405773 | 0.06 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr1_-_28241226 | 0.06 |
ENST00000373912.3
ENST00000373909.3 |
RPA2
|
replication protein A2, 32kDa |
| chr17_+_21030260 | 0.06 |
ENST00000579303.1
|
DHRS7B
|
dehydrogenase/reductase (SDR family) member 7B |
| chr17_+_7328925 | 0.06 |
ENST00000333870.3
ENST00000574034.1 |
C17orf74
|
chromosome 17 open reading frame 74 |
| chr12_-_121342170 | 0.06 |
ENST00000353487.2
|
SPPL3
|
signal peptide peptidase like 3 |
| chr7_+_80275953 | 0.06 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chrX_-_110655306 | 0.05 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr3_-_12587055 | 0.05 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr12_-_95510743 | 0.05 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr9_+_130565147 | 0.05 |
ENST00000373247.2
ENST00000373245.1 ENST00000393706.2 ENST00000373228.1 |
FPGS
|
folylpolyglutamate synthase |
| chr17_-_40829026 | 0.05 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr3_-_71353892 | 0.05 |
ENST00000484350.1
|
FOXP1
|
forkhead box P1 |
| chr12_+_12870055 | 0.05 |
ENST00000228872.4
|
CDKN1B
|
cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
| chr3_+_179280668 | 0.05 |
ENST00000429709.2
ENST00000450518.2 ENST00000392662.1 ENST00000490364.1 |
ACTL6A
|
actin-like 6A |
| chr13_+_74805561 | 0.05 |
ENST00000419499.1
|
LINC00402
|
long intergenic non-protein coding RNA 402 |
| chr4_-_186570679 | 0.05 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr14_+_104182061 | 0.05 |
ENST00000216602.6
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr16_-_29499154 | 0.05 |
ENST00000354563.5
|
RP11-231C14.4
|
Uncharacterized protein |
| chr11_+_66384053 | 0.05 |
ENST00000310137.4
ENST00000393979.3 ENST00000409372.1 ENST00000443702.1 ENST00000409738.4 ENST00000514361.3 ENST00000503028.2 ENST00000412278.2 ENST00000500635.2 |
RBM14
RBM4
RBM14-RBM4
|
RNA binding motif protein 14 RNA binding motif protein 4 RBM14-RBM4 readthrough |
| chr12_-_56615485 | 0.05 |
ENST00000549038.1
ENST00000552244.1 |
RNF41
|
ring finger protein 41 |
| chr8_+_98788057 | 0.05 |
ENST00000517924.1
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr14_-_74551172 | 0.05 |
ENST00000553458.1
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chrX_-_92928557 | 0.05 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr18_-_48346415 | 0.05 |
ENST00000431965.2
ENST00000436348.2 |
MRO
|
maestro |
| chr7_+_80275752 | 0.05 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr17_+_73663402 | 0.05 |
ENST00000355423.3
|
SAP30BP
|
SAP30 binding protein |
| chr12_-_56615693 | 0.05 |
ENST00000394013.2
ENST00000345093.4 ENST00000551711.1 ENST00000552656.1 |
RNF41
|
ring finger protein 41 |
| chr14_+_29236269 | 0.05 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr19_+_14640372 | 0.05 |
ENST00000215567.5
ENST00000598298.1 ENST00000596073.1 ENST00000600083.1 ENST00000436007.2 |
TECR
|
trans-2,3-enoyl-CoA reductase |
| chrX_+_55649833 | 0.05 |
ENST00000339140.3
|
FOXR2
|
forkhead box R2 |
| chr15_-_54025300 | 0.05 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr19_-_11266471 | 0.05 |
ENST00000592540.1
|
SPC24
|
SPC24, NDC80 kinetochore complex component |
| chr15_-_60695071 | 0.05 |
ENST00000557904.1
|
ANXA2
|
annexin A2 |
| chr16_+_19222479 | 0.04 |
ENST00000568433.1
|
SYT17
|
synaptotagmin XVII |
| chr5_+_140207536 | 0.04 |
ENST00000529310.1
ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr16_-_69788816 | 0.04 |
ENST00000268802.5
|
NOB1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr12_+_101988627 | 0.04 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr3_-_4927447 | 0.04 |
ENST00000449914.1
|
AC018816.3
|
Uncharacterized protein |
| chr6_-_26199471 | 0.04 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr11_+_44117099 | 0.04 |
ENST00000533608.1
|
EXT2
|
exostosin glycosyltransferase 2 |
| chr4_-_184241927 | 0.04 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr11_+_47279155 | 0.04 |
ENST00000444396.1
ENST00000457932.1 ENST00000412937.1 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr14_+_24584372 | 0.04 |
ENST00000559396.1
ENST00000558638.1 ENST00000561041.1 ENST00000559288.1 ENST00000558408.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr3_+_122785895 | 0.04 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr17_+_7758374 | 0.04 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr4_-_6557336 | 0.04 |
ENST00000507294.1
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr12_-_109221160 | 0.04 |
ENST00000326470.5
|
SSH1
|
slingshot protein phosphatase 1 |
| chr5_+_151151471 | 0.04 |
ENST00000394123.3
ENST00000543466.1 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr12_+_101988774 | 0.04 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr19_-_44306590 | 0.04 |
ENST00000377950.3
|
LYPD5
|
LY6/PLAUR domain containing 5 |
| chr10_-_91102410 | 0.04 |
ENST00000282673.4
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase |
| chr1_-_173176452 | 0.04 |
ENST00000281834.3
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr7_+_20370746 | 0.04 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr22_+_31795509 | 0.04 |
ENST00000331457.4
|
DRG1
|
developmentally regulated GTP binding protein 1 |
| chr4_-_73434498 | 0.04 |
ENST00000286657.4
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr8_-_6115044 | 0.04 |
ENST00000519555.1
|
RP11-124B13.1
|
RP11-124B13.1 |
| chr2_-_242626127 | 0.04 |
ENST00000445261.1
|
DTYMK
|
deoxythymidylate kinase (thymidylate kinase) |
| chr7_+_75024903 | 0.03 |
ENST00000323819.3
ENST00000430211.1 |
TRIM73
|
tripartite motif containing 73 |
| chr19_-_39881777 | 0.03 |
ENST00000595564.1
ENST00000221265.3 |
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr19_-_14640005 | 0.03 |
ENST00000596853.1
ENST00000596075.1 ENST00000595992.1 ENST00000396969.4 ENST00000601533.1 ENST00000598692.1 |
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr6_+_151561506 | 0.03 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr8_-_116681221 | 0.03 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr2_-_96971232 | 0.03 |
ENST00000323853.5
|
SNRNP200
|
small nuclear ribonucleoprotein 200kDa (U5) |
| chr3_+_129159039 | 0.03 |
ENST00000507564.1
ENST00000431818.2 ENST00000504021.1 ENST00000349441.2 ENST00000348417.2 ENST00000440957.2 |
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr9_+_130565487 | 0.03 |
ENST00000373225.3
ENST00000431857.1 |
FPGS
|
folylpolyglutamate synthase |
| chr2_+_79412357 | 0.03 |
ENST00000466387.1
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2 |
| chr8_+_7801144 | 0.03 |
ENST00000443676.1
|
ZNF705B
|
zinc finger protein 705B |
| chrM_+_14741 | 0.03 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr12_+_123944070 | 0.03 |
ENST00000412157.2
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr6_+_36839616 | 0.03 |
ENST00000359359.2
ENST00000510325.2 |
C6orf89
|
chromosome 6 open reading frame 89 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PBX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.3 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.1 | 0.3 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 0.3 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.2 | GO:2000974 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.1 | 0.2 | GO:0021569 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.2 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0052362 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.2 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0072025 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.2 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.0 | 0.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.2 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.0 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.0 | GO:2000525 | negative regulation of regulatory T cell differentiation(GO:0045590) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.1 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.0 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |