Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
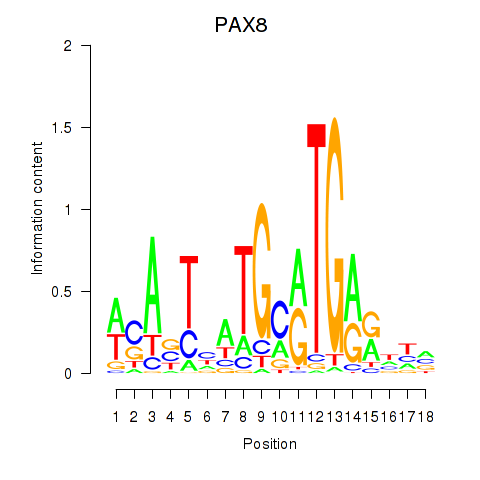
Results for PAX8
Z-value: 0.84
Transcription factors associated with PAX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX8
|
ENSG00000125618.12 | paired box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX8 | hg19_v2_chr2_-_114036488_114036527 | 0.61 | 2.0e-01 | Click! |
Activity profile of PAX8 motif
Sorted Z-values of PAX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_92821922 | 1.08 |
ENST00000538965.1
ENST00000378487.2 |
CLLU1OS
|
chronic lymphocytic leukemia up-regulated 1 opposite strand |
| chr1_-_153013588 | 1.06 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chrX_-_153141783 | 0.77 |
ENST00000458029.1
|
L1CAM
|
L1 cell adhesion molecule |
| chr12_+_72058130 | 0.64 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr2_-_37068530 | 0.62 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr11_-_105010320 | 0.43 |
ENST00000532895.1
ENST00000530950.1 |
CARD18
|
caspase recruitment domain family, member 18 |
| chr6_+_8652370 | 0.43 |
ENST00000503668.1
|
HULC
|
hepatocellular carcinoma up-regulated long non-coding RNA |
| chr10_+_12391685 | 0.42 |
ENST00000378845.1
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr14_-_57197224 | 0.38 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr1_+_218683438 | 0.37 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr12_+_7167980 | 0.37 |
ENST00000360817.5
ENST00000402681.3 |
C1S
|
complement component 1, s subcomponent |
| chr3_-_110612323 | 0.34 |
ENST00000383686.2
|
RP11-553A10.1
|
Uncharacterized protein |
| chrX_-_84363974 | 0.32 |
ENST00000395409.3
ENST00000332921.5 ENST00000509231.1 |
SATL1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr10_+_135340859 | 0.32 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr9_-_117853297 | 0.30 |
ENST00000542877.1
ENST00000537320.1 ENST00000341037.4 |
TNC
|
tenascin C |
| chr6_-_6711235 | 0.29 |
ENST00000432823.2
|
RP1-80N2.2
|
RP1-80N2.2 |
| chr4_+_123653882 | 0.29 |
ENST00000433287.1
|
BBS12
|
Bardet-Biedl syndrome 12 |
| chr1_-_52520828 | 0.28 |
ENST00000610127.1
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr17_-_73267214 | 0.28 |
ENST00000580717.1
ENST00000577542.1 ENST00000579612.1 ENST00000245551.5 |
MIF4GD
|
MIF4G domain containing |
| chr22_-_44258280 | 0.28 |
ENST00000540422.1
|
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr11_-_9482010 | 0.26 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr17_+_7462103 | 0.26 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr6_+_24126350 | 0.26 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr17_-_73267304 | 0.25 |
ENST00000579297.1
ENST00000580571.1 |
MIF4GD
|
MIF4G domain containing |
| chr17_-_73267281 | 0.24 |
ENST00000578305.1
ENST00000325102.8 |
MIF4GD
|
MIF4G domain containing |
| chr6_-_44233361 | 0.24 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr19_+_34891252 | 0.24 |
ENST00000606020.1
|
RP11-618P17.4
|
Uncharacterized protein |
| chr1_-_119682812 | 0.23 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr17_+_7461613 | 0.23 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr19_+_35417939 | 0.23 |
ENST00000601142.1
ENST00000426813.2 |
ZNF30
|
zinc finger protein 30 |
| chr19_+_42041860 | 0.22 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr19_-_55791563 | 0.21 |
ENST00000588971.1
ENST00000255631.5 ENST00000587551.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr17_+_7461580 | 0.21 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr12_-_123201337 | 0.21 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr11_-_102401469 | 0.21 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr2_-_106013207 | 0.20 |
ENST00000447958.1
|
FHL2
|
four and a half LIM domains 2 |
| chr20_-_36794938 | 0.20 |
ENST00000453095.1
|
TGM2
|
transglutaminase 2 |
| chr13_-_24471194 | 0.20 |
ENST00000382137.3
ENST00000382057.3 |
C1QTNF9B
|
C1q and tumor necrosis factor related protein 9B |
| chr19_+_782755 | 0.19 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr12_-_86650077 | 0.19 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr15_+_67420441 | 0.19 |
ENST00000558894.1
|
SMAD3
|
SMAD family member 3 |
| chr6_+_43739697 | 0.19 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr6_-_134639180 | 0.18 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr21_-_44582378 | 0.18 |
ENST00000433840.1
|
AP001631.10
|
AP001631.10 |
| chr13_+_31309645 | 0.18 |
ENST00000380490.3
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein |
| chr1_-_182573514 | 0.17 |
ENST00000367558.5
|
RGS16
|
regulator of G-protein signaling 16 |
| chr7_+_129984630 | 0.17 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr14_+_75230011 | 0.17 |
ENST00000552421.1
ENST00000325680.7 ENST00000238571.3 |
YLPM1
|
YLP motif containing 1 |
| chr11_-_2170786 | 0.17 |
ENST00000300632.5
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr11_+_59705928 | 0.16 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr8_-_121825088 | 0.16 |
ENST00000520717.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr6_+_161123270 | 0.16 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr8_+_86851932 | 0.16 |
ENST00000517368.1
|
CTA-392E5.1
|
CTA-392E5.1 |
| chr4_+_123653807 | 0.16 |
ENST00000314218.3
ENST00000542236.1 |
BBS12
|
Bardet-Biedl syndrome 12 |
| chr10_+_103912137 | 0.15 |
ENST00000603742.1
ENST00000488254.2 ENST00000461421.1 ENST00000476468.1 ENST00000370007.5 |
NOLC1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr3_+_190105909 | 0.15 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr12_-_123187890 | 0.15 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr6_+_14117872 | 0.15 |
ENST00000379153.3
|
CD83
|
CD83 molecule |
| chr11_+_82904746 | 0.15 |
ENST00000393389.3
ENST00000528722.1 |
ANKRD42
|
ankyrin repeat domain 42 |
| chr3_-_161090660 | 0.15 |
ENST00000359175.4
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr2_+_185463093 | 0.15 |
ENST00000302277.6
|
ZNF804A
|
zinc finger protein 804A |
| chr1_+_41448820 | 0.15 |
ENST00000372616.1
|
CTPS1
|
CTP synthase 1 |
| chr9_+_42704004 | 0.14 |
ENST00000457288.1
|
CBWD7
|
COBW domain containing 7 |
| chrX_+_129535937 | 0.14 |
ENST00000305536.6
ENST00000370947.1 |
RBMX2
|
RNA binding motif protein, X-linked 2 |
| chr15_-_89010607 | 0.14 |
ENST00000312475.4
|
MRPL46
|
mitochondrial ribosomal protein L46 |
| chr6_+_135502408 | 0.14 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr11_-_47870091 | 0.14 |
ENST00000526870.1
|
NUP160
|
nucleoporin 160kDa |
| chr17_-_72588422 | 0.14 |
ENST00000375352.1
|
CD300LD
|
CD300 molecule-like family member d |
| chr12_-_56236690 | 0.14 |
ENST00000322569.4
|
MMP19
|
matrix metallopeptidase 19 |
| chr17_-_34257731 | 0.13 |
ENST00000431884.2
ENST00000425909.3 ENST00000394528.3 ENST00000430160.2 |
RDM1
|
RAD52 motif 1 |
| chr22_-_31324215 | 0.13 |
ENST00000429468.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr17_-_73266616 | 0.13 |
ENST00000579194.1
ENST00000581777.1 |
MIF4GD
|
MIF4G domain containing |
| chrM_+_4431 | 0.13 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr1_+_186649754 | 0.13 |
ENST00000608917.1
|
RP5-973M2.2
|
RP5-973M2.2 |
| chr7_+_6713376 | 0.13 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr7_-_76255444 | 0.12 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr1_+_100817262 | 0.12 |
ENST00000455467.1
|
CDC14A
|
cell division cycle 14A |
| chr1_+_207038699 | 0.12 |
ENST00000367098.1
|
IL20
|
interleukin 20 |
| chr19_+_42301079 | 0.12 |
ENST00000596544.1
|
CEACAM3
|
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr17_+_6918093 | 0.12 |
ENST00000439424.2
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr5_-_176943917 | 0.12 |
ENST00000330503.7
|
DDX41
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 |
| chr4_-_144826682 | 0.12 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr8_+_31496809 | 0.12 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr11_-_62609281 | 0.12 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr8_-_66474884 | 0.11 |
ENST00000520902.1
|
CTD-3025N20.2
|
CTD-3025N20.2 |
| chr7_-_144533074 | 0.11 |
ENST00000360057.3
ENST00000378099.3 |
TPK1
|
thiamin pyrophosphokinase 1 |
| chr1_-_186649543 | 0.11 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr17_-_39216344 | 0.11 |
ENST00000391418.2
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr19_-_51538148 | 0.11 |
ENST00000319590.4
ENST00000250351.4 |
KLK12
|
kallikrein-related peptidase 12 |
| chr12_-_56236734 | 0.11 |
ENST00000548629.1
|
MMP19
|
matrix metallopeptidase 19 |
| chr14_-_75083313 | 0.11 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr17_-_1508379 | 0.11 |
ENST00000412517.3
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr2_+_90248739 | 0.11 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr19_+_18794470 | 0.11 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr11_-_58343319 | 0.11 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chr14_-_91710852 | 0.10 |
ENST00000535815.1
ENST00000529102.1 |
GPR68
|
G protein-coupled receptor 68 |
| chr7_+_149535455 | 0.10 |
ENST00000223210.4
ENST00000460379.1 |
ZNF862
|
zinc finger protein 862 |
| chr5_+_69321074 | 0.10 |
ENST00000380751.5
ENST00000380750.3 ENST00000503931.1 ENST00000506542.1 |
SERF1B
|
small EDRK-rich factor 1B (centromeric) |
| chr19_-_52254050 | 0.10 |
ENST00000600815.1
|
FPR1
|
formyl peptide receptor 1 |
| chr20_+_4152356 | 0.10 |
ENST00000379460.2
|
SMOX
|
spermine oxidase |
| chr16_-_31519691 | 0.10 |
ENST00000567994.1
ENST00000430477.2 ENST00000570164.1 ENST00000327237.2 |
C16orf58
|
chromosome 16 open reading frame 58 |
| chr17_-_73781567 | 0.10 |
ENST00000586607.1
|
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr22_+_38082330 | 0.10 |
ENST00000359114.4
|
NOL12
|
nucleolar protein 12 |
| chr17_+_9745786 | 0.09 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr17_+_7461849 | 0.09 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr11_-_74022658 | 0.09 |
ENST00000427714.2
ENST00000331597.4 |
P4HA3
|
prolyl 4-hydroxylase, alpha polypeptide III |
| chr4_+_123747834 | 0.09 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr21_-_27462351 | 0.09 |
ENST00000448850.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr1_-_71513471 | 0.09 |
ENST00000370931.3
ENST00000356595.4 ENST00000306666.5 ENST00000370932.2 ENST00000351052.5 ENST00000414819.1 ENST00000370924.4 |
PTGER3
|
prostaglandin E receptor 3 (subtype EP3) |
| chr19_+_40476912 | 0.09 |
ENST00000157812.2
|
PSMC4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr14_+_66578299 | 0.09 |
ENST00000554187.1
ENST00000556662.1 ENST00000556291.1 ENST00000557723.1 ENST00000557050.1 |
RP11-783L4.1
|
RP11-783L4.1 |
| chr19_-_6057282 | 0.09 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chrY_-_20935572 | 0.09 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chr19_+_42300548 | 0.09 |
ENST00000344550.4
|
CEACAM3
|
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr14_-_54425475 | 0.09 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr6_+_30844192 | 0.09 |
ENST00000502955.1
ENST00000505066.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr6_+_160542870 | 0.09 |
ENST00000324965.4
ENST00000457470.2 |
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr12_-_6960407 | 0.08 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr19_-_51537982 | 0.08 |
ENST00000525263.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr3_-_127842612 | 0.08 |
ENST00000417360.1
ENST00000322623.5 |
RUVBL1
|
RuvB-like AAA ATPase 1 |
| chr12_+_53689309 | 0.08 |
ENST00000351500.3
ENST00000550846.1 ENST00000334478.4 ENST00000549759.1 |
PFDN5
|
prefoldin subunit 5 |
| chr3_-_15540055 | 0.08 |
ENST00000605797.1
ENST00000435459.2 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr8_+_27168988 | 0.08 |
ENST00000397501.1
ENST00000338238.4 ENST00000544172.1 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr7_+_95115210 | 0.08 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr11_+_117857063 | 0.08 |
ENST00000227752.3
ENST00000541785.1 ENST00000545409.1 |
IL10RA
|
interleukin 10 receptor, alpha |
| chr11_-_129062093 | 0.08 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr3_-_110612059 | 0.08 |
ENST00000485473.1
|
RP11-553A10.1
|
Uncharacterized protein |
| chr8_-_7220490 | 0.08 |
ENST00000400078.2
|
ZNF705G
|
zinc finger protein 705G |
| chr4_+_36283213 | 0.08 |
ENST00000357504.3
|
DTHD1
|
death domain containing 1 |
| chr7_+_75677354 | 0.08 |
ENST00000461263.2
ENST00000315758.5 ENST00000443006.1 |
MDH2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr12_+_57828521 | 0.08 |
ENST00000309668.2
|
INHBC
|
inhibin, beta C |
| chr12_-_56236711 | 0.08 |
ENST00000409200.3
|
MMP19
|
matrix metallopeptidase 19 |
| chrY_+_20708557 | 0.08 |
ENST00000307393.2
ENST00000309834.4 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor, Y-linked 1 |
| chr13_+_76362974 | 0.08 |
ENST00000497947.2
|
LMO7
|
LIM domain 7 |
| chr11_-_47870019 | 0.08 |
ENST00000378460.2
|
NUP160
|
nucleoporin 160kDa |
| chr1_-_119683251 | 0.08 |
ENST00000369426.5
ENST00000235521.4 |
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr3_-_107941209 | 0.08 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr15_-_60683326 | 0.08 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr4_-_23735183 | 0.08 |
ENST00000507666.1
|
RP11-380P13.2
|
RP11-380P13.2 |
| chr19_-_37701386 | 0.08 |
ENST00000527838.1
ENST00000591492.1 ENST00000532828.2 |
ZNF585B
|
zinc finger protein 585B |
| chr17_+_38444115 | 0.08 |
ENST00000580824.1
ENST00000577249.1 |
CDC6
|
cell division cycle 6 |
| chr11_-_18063973 | 0.07 |
ENST00000528338.1
|
TPH1
|
tryptophan hydroxylase 1 |
| chr19_-_55791540 | 0.07 |
ENST00000433386.2
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr1_-_25256368 | 0.07 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chr4_+_71600144 | 0.07 |
ENST00000502653.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr16_+_66413128 | 0.07 |
ENST00000563425.2
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr16_-_87350970 | 0.07 |
ENST00000567970.1
|
C16orf95
|
chromosome 16 open reading frame 95 |
| chr19_+_9938562 | 0.07 |
ENST00000586895.1
ENST00000358666.3 ENST00000590068.1 ENST00000593087.1 |
UBL5
|
ubiquitin-like 5 |
| chr17_-_8301132 | 0.07 |
ENST00000399398.2
|
RNF222
|
ring finger protein 222 |
| chr19_-_44860820 | 0.07 |
ENST00000354340.4
ENST00000337401.4 ENST00000587909.1 |
ZNF112
|
zinc finger protein 112 |
| chr9_-_92020841 | 0.07 |
ENST00000433650.1
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr4_+_75230853 | 0.07 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr6_+_135502466 | 0.07 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr4_+_119809984 | 0.06 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr7_-_47520869 | 0.06 |
ENST00000415929.1
|
TNS3
|
tensin 3 |
| chr19_+_41856816 | 0.06 |
ENST00000539627.1
|
TMEM91
|
transmembrane protein 91 |
| chr15_+_45021183 | 0.06 |
ENST00000559390.1
|
TRIM69
|
tripartite motif containing 69 |
| chr9_+_118950325 | 0.06 |
ENST00000534838.1
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr11_+_60609537 | 0.06 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr17_-_67057047 | 0.06 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr6_+_135502501 | 0.06 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr12_-_129308487 | 0.06 |
ENST00000266771.5
|
SLC15A4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr6_+_36853607 | 0.06 |
ENST00000480824.2
ENST00000355190.3 ENST00000373685.1 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr13_-_103426112 | 0.06 |
ENST00000376032.4
ENST00000376029.3 |
TEX30
|
testis expressed 30 |
| chr16_-_30773372 | 0.06 |
ENST00000545825.1
ENST00000541260.1 |
C16orf93
|
chromosome 16 open reading frame 93 |
| chr1_-_8585945 | 0.06 |
ENST00000377464.1
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr11_+_9482512 | 0.06 |
ENST00000396602.2
ENST00000530463.1 ENST00000533542.1 ENST00000532577.1 ENST00000396597.3 |
ZNF143
|
zinc finger protein 143 |
| chr3_-_112329110 | 0.06 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr5_+_140027355 | 0.06 |
ENST00000417647.2
ENST00000507593.1 ENST00000508301.1 |
IK
|
IK cytokine, down-regulator of HLA II |
| chr5_-_133510456 | 0.06 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr13_-_25746416 | 0.05 |
ENST00000515384.1
ENST00000357816.2 |
AMER2
|
APC membrane recruitment protein 2 |
| chr11_-_8816375 | 0.05 |
ENST00000530580.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr17_-_27278304 | 0.05 |
ENST00000577226.1
|
PHF12
|
PHD finger protein 12 |
| chr10_+_24738355 | 0.05 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr2_-_132589601 | 0.05 |
ENST00000437330.1
|
AC103564.7
|
AC103564.7 |
| chr1_+_180165672 | 0.05 |
ENST00000443059.1
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr9_+_27109392 | 0.05 |
ENST00000406359.4
|
TEK
|
TEK tyrosine kinase, endothelial |
| chr17_+_7461781 | 0.05 |
ENST00000349228.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chrX_-_153141302 | 0.05 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr3_+_150264458 | 0.05 |
ENST00000487799.1
ENST00000460851.1 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr11_+_20044375 | 0.05 |
ENST00000525322.1
ENST00000530408.1 |
NAV2
|
neuron navigator 2 |
| chr8_-_53322303 | 0.05 |
ENST00000276480.7
|
ST18
|
suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) |
| chr19_+_16296191 | 0.05 |
ENST00000589852.1
ENST00000263384.7 ENST00000588367.1 ENST00000587351.1 |
FAM32A
|
family with sequence similarity 32, member A |
| chr4_-_16077741 | 0.05 |
ENST00000447510.2
ENST00000540805.1 ENST00000539194.1 |
PROM1
|
prominin 1 |
| chr12_+_122516626 | 0.05 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chr6_-_31628512 | 0.05 |
ENST00000375911.1
|
C6orf47
|
chromosome 6 open reading frame 47 |
| chr18_-_29264467 | 0.05 |
ENST00000383131.3
ENST00000237019.7 |
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr20_-_5931109 | 0.05 |
ENST00000203001.2
|
TRMT6
|
tRNA methyltransferase 6 homolog (S. cerevisiae) |
| chr5_+_140235469 | 0.05 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr17_-_33760269 | 0.05 |
ENST00000452764.3
|
SLFN12
|
schlafen family member 12 |
| chr10_-_5638048 | 0.05 |
ENST00000478294.1
|
RP13-463N16.6
|
RP13-463N16.6 |
| chr13_-_95131923 | 0.05 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr8_-_53322347 | 0.05 |
ENST00000517580.1
|
ST18
|
suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) |
| chr12_-_10251603 | 0.04 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr19_+_29097595 | 0.04 |
ENST00000587961.1
|
AC079466.1
|
AC079466.1 |
| chr4_-_10686475 | 0.04 |
ENST00000226951.6
|
CLNK
|
cytokine-dependent hematopoietic cell linker |
| chr3_-_8811288 | 0.04 |
ENST00000316793.3
ENST00000431493.1 |
OXTR
|
oxytocin receptor |
| chr19_-_51538118 | 0.04 |
ENST00000529888.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr15_+_34638066 | 0.04 |
ENST00000333756.4
|
NUTM1
|
NUT midline carcinoma, family member 1 |
| chr2_-_175712479 | 0.04 |
ENST00000443238.1
|
CHN1
|
chimerin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.3 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.3 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.9 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.3 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.2 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.2 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.3 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 0.2 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.2 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 0.0 | 0.3 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.4 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0052417 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.1 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.2 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:2000007 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 1.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.4 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.2 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0097449 | astrocyte projection(GO:0097449) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.1 | 0.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.2 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.1 | 0.3 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.1 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.0 | 0.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.3 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.3 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |