Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
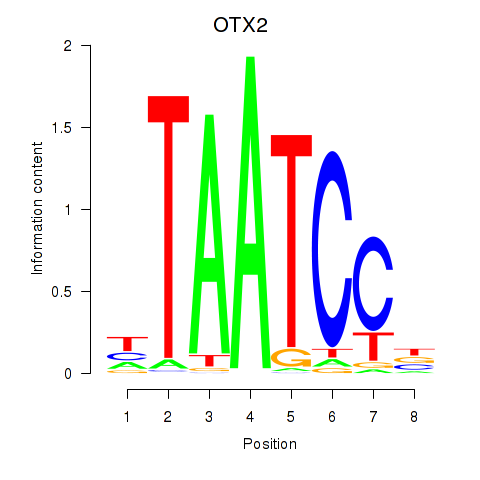
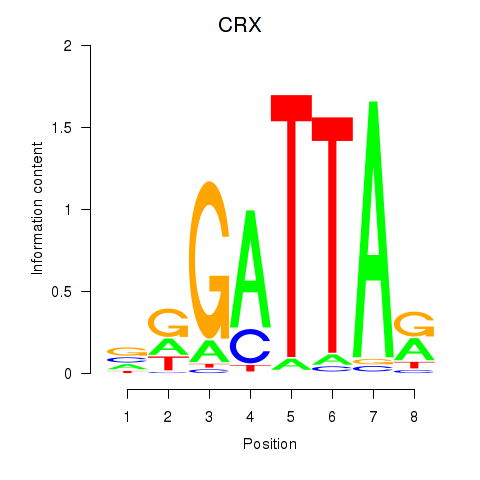
Results for OTX2_CRX
Z-value: 0.50
Transcription factors associated with OTX2_CRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTX2
|
ENSG00000165588.12 | orthodenticle homeobox 2 |
|
CRX
|
ENSG00000105392.11 | cone-rod homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CRX | hg19_v2_chr19_+_48325097_48325211 | 0.23 | 6.7e-01 | Click! |
Activity profile of OTX2_CRX motif
Sorted Z-values of OTX2_CRX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_44092784 | 0.45 |
ENST00000458412.1
|
HYPK
|
huntingtin interacting protein K |
| chrX_+_69488174 | 0.31 |
ENST00000480877.2
ENST00000307959.8 |
ARR3
|
arrestin 3, retinal (X-arrestin) |
| chr7_-_100171270 | 0.27 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr17_+_74536164 | 0.25 |
ENST00000586148.1
|
PRCD
|
progressive rod-cone degeneration |
| chr17_-_38859996 | 0.25 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr17_-_40264692 | 0.24 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chrX_+_69488155 | 0.22 |
ENST00000374495.3
|
ARR3
|
arrestin 3, retinal (X-arrestin) |
| chr16_-_31076332 | 0.20 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr15_-_74494779 | 0.19 |
ENST00000571341.1
|
STRA6
|
stimulated by retinoic acid 6 |
| chr5_+_133451254 | 0.18 |
ENST00000517851.1
ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr7_-_128415844 | 0.17 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr19_-_51538148 | 0.17 |
ENST00000319590.4
ENST00000250351.4 |
KLK12
|
kallikrein-related peptidase 12 |
| chrX_+_15767971 | 0.17 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr5_+_5140436 | 0.16 |
ENST00000511368.1
ENST00000274181.7 |
ADAMTS16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16 |
| chr11_-_36619771 | 0.16 |
ENST00000311485.3
ENST00000527033.1 ENST00000532616.1 |
RAG2
|
recombination activating gene 2 |
| chr8_-_25281747 | 0.16 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr12_+_85673868 | 0.15 |
ENST00000316824.3
|
ALX1
|
ALX homeobox 1 |
| chr18_+_32290218 | 0.15 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr7_-_17500294 | 0.13 |
ENST00000439046.1
|
AC019117.2
|
AC019117.2 |
| chrX_+_153769446 | 0.13 |
ENST00000422680.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr19_-_51538118 | 0.13 |
ENST00000529888.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr6_+_148593425 | 0.12 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr14_-_22005197 | 0.12 |
ENST00000541965.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr9_-_98268883 | 0.11 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr1_-_165414414 | 0.11 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr6_-_33297013 | 0.11 |
ENST00000453407.1
|
DAXX
|
death-domain associated protein |
| chr19_-_55866104 | 0.11 |
ENST00000326529.4
|
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chrX_-_15619076 | 0.11 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr7_-_97881429 | 0.10 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr6_+_35310312 | 0.10 |
ENST00000448077.2
ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr3_+_49058444 | 0.09 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr16_-_31076273 | 0.09 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chr1_+_200863949 | 0.09 |
ENST00000413687.2
|
C1orf106
|
chromosome 1 open reading frame 106 |
| chr17_-_39661947 | 0.09 |
ENST00000590425.1
|
KRT13
|
keratin 13 |
| chr5_-_88179302 | 0.09 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr10_+_103986085 | 0.08 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr17_-_4643161 | 0.08 |
ENST00000574412.1
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr17_-_43210580 | 0.08 |
ENST00000538093.1
ENST00000590644.1 |
PLCD3
|
phospholipase C, delta 3 |
| chr6_+_35310391 | 0.08 |
ENST00000337400.2
ENST00000311565.4 ENST00000540939.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr15_-_74495188 | 0.08 |
ENST00000563965.1
ENST00000395105.4 |
STRA6
|
stimulated by retinoic acid 6 |
| chr19_-_55866061 | 0.07 |
ENST00000588572.2
ENST00000593184.1 ENST00000589467.1 |
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr1_-_26633480 | 0.07 |
ENST00000450041.1
|
UBXN11
|
UBX domain protein 11 |
| chr3_+_49057876 | 0.07 |
ENST00000326912.4
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr11_+_72975524 | 0.07 |
ENST00000540342.1
ENST00000542092.1 |
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr2_-_14541060 | 0.07 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr1_-_117664317 | 0.07 |
ENST00000256649.4
ENST00000369464.3 ENST00000485032.1 |
TRIM45
|
tripartite motif containing 45 |
| chr4_+_124571409 | 0.07 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr2_+_183943464 | 0.06 |
ENST00000354221.4
|
DUSP19
|
dual specificity phosphatase 19 |
| chr5_-_41794313 | 0.06 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr3_-_49058479 | 0.06 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr11_+_72975559 | 0.06 |
ENST00000349767.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr17_-_39661849 | 0.06 |
ENST00000246635.3
ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr16_+_58010339 | 0.06 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr5_-_149324306 | 0.06 |
ENST00000255266.5
|
PDE6A
|
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| chr10_-_94003003 | 0.06 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chrX_-_124097620 | 0.06 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr19_+_48325097 | 0.05 |
ENST00000221996.7
ENST00000539067.1 |
CRX
|
cone-rod homeobox |
| chr18_-_70532906 | 0.05 |
ENST00000299430.2
ENST00000397929.1 |
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr5_+_50679506 | 0.05 |
ENST00000511384.1
|
ISL1
|
ISL LIM homeobox 1 |
| chr6_+_130339710 | 0.05 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr2_-_175711133 | 0.05 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr19_-_51537982 | 0.05 |
ENST00000525263.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chrX_-_153363125 | 0.05 |
ENST00000407218.1
ENST00000453960.2 |
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr5_-_88179017 | 0.05 |
ENST00000514028.1
ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr20_-_18774614 | 0.05 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr3_+_49059038 | 0.05 |
ENST00000451378.2
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr11_+_101983176 | 0.05 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chrX_+_153769409 | 0.05 |
ENST00000440286.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr14_+_96722152 | 0.05 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr16_-_58004992 | 0.05 |
ENST00000564448.1
ENST00000251102.8 ENST00000311183.4 |
CNGB1
|
cyclic nucleotide gated channel beta 1 |
| chr9_-_127358087 | 0.05 |
ENST00000475178.1
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr1_+_164528437 | 0.04 |
ENST00000485769.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr19_-_42931567 | 0.04 |
ENST00000244289.4
|
LIPE
|
lipase, hormone-sensitive |
| chr11_+_72975578 | 0.04 |
ENST00000393592.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr17_-_4689727 | 0.04 |
ENST00000328739.5
ENST00000354194.4 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr17_+_16945820 | 0.04 |
ENST00000577514.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chrX_-_49089771 | 0.04 |
ENST00000376251.1
ENST00000323022.5 ENST00000376265.2 |
CACNA1F
|
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr18_-_64271363 | 0.04 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr14_-_71107921 | 0.04 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr9_-_131486367 | 0.04 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr5_-_524445 | 0.04 |
ENST00000514375.1
ENST00000264938.3 |
SLC9A3
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 |
| chr1_+_180875711 | 0.04 |
ENST00000434447.1
|
RP11-46A10.2
|
RP11-46A10.2 |
| chr7_-_128171123 | 0.03 |
ENST00000608477.1
|
RP11-212P7.2
|
RP11-212P7.2 |
| chr20_-_31124186 | 0.03 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr13_-_49975632 | 0.03 |
ENST00000457041.1
ENST00000355854.4 |
CAB39L
|
calcium binding protein 39-like |
| chr17_+_43318434 | 0.03 |
ENST00000587489.1
|
FMNL1
|
formin-like 1 |
| chr14_-_101034407 | 0.03 |
ENST00000443071.2
ENST00000557378.1 |
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr7_-_78400598 | 0.03 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr5_-_88120083 | 0.03 |
ENST00000509373.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr3_+_50649302 | 0.03 |
ENST00000446044.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr1_+_11751748 | 0.03 |
ENST00000294485.5
|
DRAXIN
|
dorsal inhibitory axon guidance protein |
| chrX_+_28605516 | 0.03 |
ENST00000378993.1
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1 |
| chr14_-_22005343 | 0.03 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr1_-_186430222 | 0.03 |
ENST00000391997.2
|
PDC
|
phosducin |
| chr22_-_41258074 | 0.03 |
ENST00000307221.4
|
DNAJB7
|
DnaJ (Hsp40) homolog, subfamily B, member 7 |
| chr15_+_63050785 | 0.03 |
ENST00000472902.1
|
TLN2
|
talin 2 |
| chr2_-_225362533 | 0.03 |
ENST00000451538.1
|
CUL3
|
cullin 3 |
| chr12_+_81101277 | 0.03 |
ENST00000228641.3
|
MYF6
|
myogenic factor 6 (herculin) |
| chr3_-_9291063 | 0.03 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr19_-_3479086 | 0.03 |
ENST00000587847.1
|
C19orf77
|
chromosome 19 open reading frame 77 |
| chr19_+_56166360 | 0.03 |
ENST00000308924.4
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2 |
| chr6_-_42690312 | 0.03 |
ENST00000230381.5
|
PRPH2
|
peripherin 2 (retinal degeneration, slow) |
| chr19_+_11457175 | 0.03 |
ENST00000458408.1
ENST00000586451.1 ENST00000588592.1 |
CCDC159
|
coiled-coil domain containing 159 |
| chr17_+_72931876 | 0.03 |
ENST00000328801.4
|
OTOP3
|
otopetrin 3 |
| chr5_-_58882219 | 0.03 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr3_-_50649192 | 0.03 |
ENST00000443053.2
ENST00000348721.3 |
CISH
|
cytokine inducible SH2-containing protein |
| chr21_+_33245548 | 0.03 |
ENST00000270112.2
|
HUNK
|
hormonally up-regulated Neu-associated kinase |
| chr11_-_8680383 | 0.03 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr12_-_86650077 | 0.03 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr19_+_18208603 | 0.03 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr9_+_34992846 | 0.03 |
ENST00000443266.1
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr10_-_74114714 | 0.03 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr9_-_139361503 | 0.03 |
ENST00000453963.1
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr22_-_30867973 | 0.03 |
ENST00000402286.1
ENST00000401751.1 ENST00000539629.1 ENST00000403066.1 ENST00000215812.4 |
SEC14L3
|
SEC14-like 3 (S. cerevisiae) |
| chr14_-_23395623 | 0.02 |
ENST00000556043.1
|
PRMT5
|
protein arginine methyltransferase 5 |
| chr19_+_7710774 | 0.02 |
ENST00000602355.1
|
STXBP2
|
syntaxin binding protein 2 |
| chrM_+_10758 | 0.02 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr3_-_190580404 | 0.02 |
ENST00000442080.1
|
GMNC
|
geminin coiled-coil domain containing |
| chr8_-_114449112 | 0.02 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr12_-_54779511 | 0.02 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr17_-_4689649 | 0.02 |
ENST00000441199.2
ENST00000416307.2 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr21_+_17443434 | 0.02 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr10_-_21806759 | 0.02 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr16_+_81272287 | 0.02 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr13_-_95131923 | 0.02 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr19_-_44124019 | 0.02 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr12_-_66275350 | 0.02 |
ENST00000536648.1
|
RP11-366L20.2
|
Uncharacterized protein |
| chrX_-_153363188 | 0.02 |
ENST00000303391.6
|
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr19_-_55865908 | 0.02 |
ENST00000590900.1
|
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr7_-_111424462 | 0.02 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr1_+_62439037 | 0.02 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr14_-_101034811 | 0.02 |
ENST00000553553.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr8_-_10512569 | 0.02 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr6_-_66417107 | 0.02 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr12_-_53171128 | 0.02 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chrX_+_135279179 | 0.02 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr21_+_17443521 | 0.02 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr14_-_67981916 | 0.02 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chr6_-_30658745 | 0.02 |
ENST00000376420.5
ENST00000376421.5 |
NRM
|
nurim (nuclear envelope membrane protein) |
| chr5_+_150040403 | 0.02 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr19_+_40877583 | 0.02 |
ENST00000596470.1
|
PLD3
|
phospholipase D family, member 3 |
| chr15_+_79603479 | 0.02 |
ENST00000424155.2
ENST00000536821.1 |
TMED3
|
transmembrane emp24 protein transport domain containing 3 |
| chrX_-_10588459 | 0.02 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr8_+_55528627 | 0.02 |
ENST00000220676.1
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr7_+_80231466 | 0.02 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr2_-_188312971 | 0.02 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr1_-_151965048 | 0.02 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10 |
| chr2_+_33661382 | 0.02 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr11_+_128563652 | 0.02 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr8_-_145701718 | 0.02 |
ENST00000377317.4
|
FOXH1
|
forkhead box H1 |
| chr4_-_68829226 | 0.02 |
ENST00000396188.2
|
TMPRSS11A
|
transmembrane protease, serine 11A |
| chr2_-_200715573 | 0.02 |
ENST00000420922.2
|
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr19_-_11457162 | 0.02 |
ENST00000590482.1
|
TMEM205
|
transmembrane protein 205 |
| chr20_+_4702548 | 0.02 |
ENST00000305817.2
|
PRND
|
prion protein 2 (dublet) |
| chr6_-_154568815 | 0.02 |
ENST00000519344.1
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr6_+_106535455 | 0.02 |
ENST00000424894.1
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr11_+_124735282 | 0.02 |
ENST00000397801.1
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr1_+_81771806 | 0.02 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr3_+_111393501 | 0.02 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr18_+_616672 | 0.02 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr10_-_21186144 | 0.02 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr3_-_47950745 | 0.02 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr5_-_33984741 | 0.02 |
ENST00000382102.3
ENST00000509381.1 ENST00000342059.3 ENST00000345083.5 |
SLC45A2
|
solute carrier family 45, member 2 |
| chr11_-_128712362 | 0.02 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr10_+_24528108 | 0.02 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr13_-_79980315 | 0.02 |
ENST00000438737.2
|
RBM26
|
RNA binding motif protein 26 |
| chr17_-_43209862 | 0.02 |
ENST00000322765.5
|
PLCD3
|
phospholipase C, delta 3 |
| chr20_-_49307897 | 0.02 |
ENST00000535356.1
|
FAM65C
|
family with sequence similarity 65, member C |
| chr4_-_68829144 | 0.02 |
ENST00000508048.1
|
TMPRSS11A
|
transmembrane protease, serine 11A |
| chr11_-_111794446 | 0.01 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr11_-_71753188 | 0.01 |
ENST00000543009.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr10_+_52750930 | 0.01 |
ENST00000401604.2
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr12_+_58138664 | 0.01 |
ENST00000257910.3
|
TSPAN31
|
tetraspanin 31 |
| chr11_-_128894053 | 0.01 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr6_-_30899924 | 0.01 |
ENST00000359086.3
|
SFTA2
|
surfactant associated 2 |
| chrM_+_8527 | 0.01 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chrX_-_33357558 | 0.01 |
ENST00000288447.4
|
DMD
|
dystrophin |
| chr17_-_60885659 | 0.01 |
ENST00000311269.5
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr11_+_20044096 | 0.01 |
ENST00000533917.1
|
NAV2
|
neuron navigator 2 |
| chr4_+_110769258 | 0.01 |
ENST00000594814.1
|
LRIT3
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 |
| chr10_-_104178857 | 0.01 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chrX_-_100129128 | 0.01 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr9_+_75229616 | 0.01 |
ENST00000340019.3
|
TMC1
|
transmembrane channel-like 1 |
| chr5_+_145718587 | 0.01 |
ENST00000230732.4
|
POU4F3
|
POU class 4 homeobox 3 |
| chr10_-_126849588 | 0.01 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr1_+_164528866 | 0.01 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr18_+_616711 | 0.01 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr7_+_136553370 | 0.01 |
ENST00000445907.2
|
CHRM2
|
cholinergic receptor, muscarinic 2 |
| chr7_-_83824169 | 0.01 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_-_60885700 | 0.01 |
ENST00000583600.1
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr1_+_209757051 | 0.01 |
ENST00000009105.1
ENST00000423146.1 ENST00000361322.2 |
CAMK1G
|
calcium/calmodulin-dependent protein kinase IG |
| chr3_+_191046810 | 0.01 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr1_+_55505184 | 0.01 |
ENST00000302118.5
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9 |
| chr19_-_59066452 | 0.01 |
ENST00000312547.2
|
CHMP2A
|
charged multivesicular body protein 2A |
| chr12_+_70133152 | 0.01 |
ENST00000550536.1
ENST00000362025.5 |
RAB3IP
|
RAB3A interacting protein |
| chr18_-_21891460 | 0.01 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr10_+_129785574 | 0.01 |
ENST00000430713.2
ENST00000471218.1 |
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr15_+_54901540 | 0.01 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr9_+_18474098 | 0.01 |
ENST00000327883.7
ENST00000431052.2 ENST00000380570.4 |
ADAMTSL1
|
ADAMTS-like 1 |
| chr1_+_202431859 | 0.01 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
Network of associatons between targets according to the STRING database.
First level regulatory network of OTX2_CRX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.3 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:0003051 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.2 | GO:0048619 | embryonic genitalia morphogenesis(GO:0030538) embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.2 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.2 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.2 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.0 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |