Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
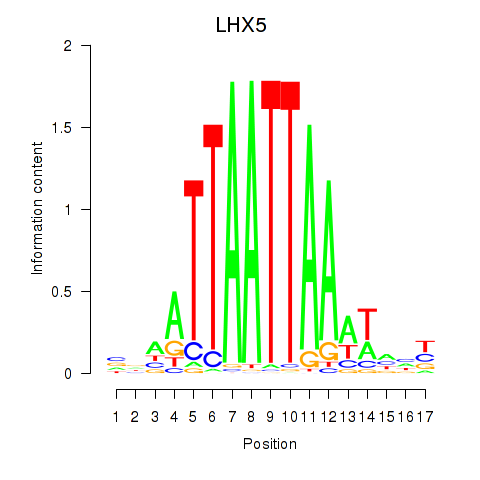
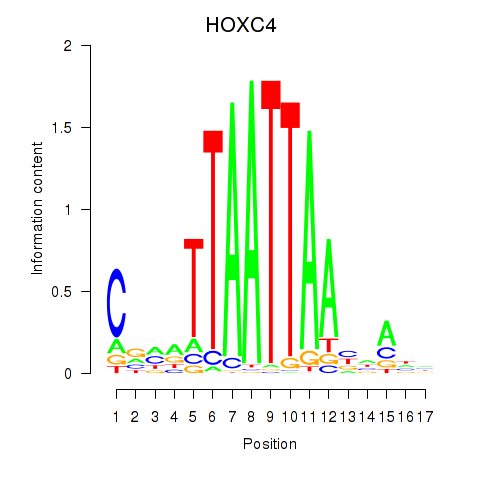
Results for OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
Z-value: 0.33
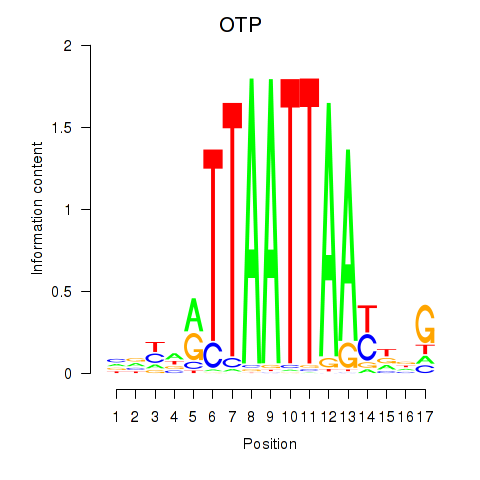
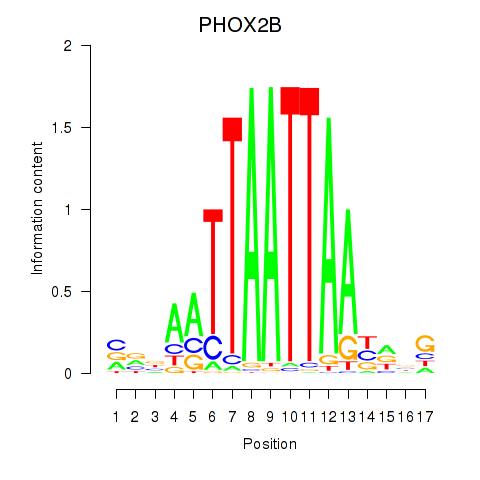
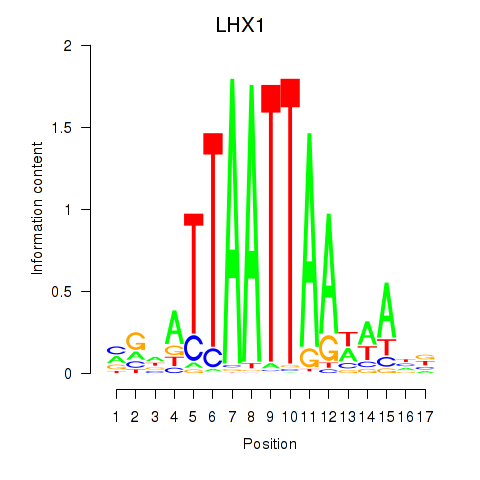
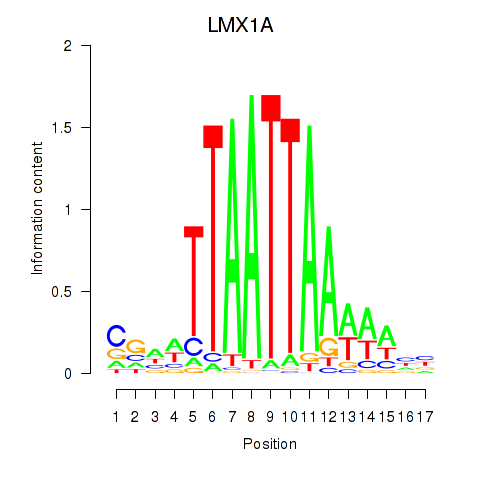
Transcription factors associated with OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTP
|
ENSG00000171540.6 | orthopedia homeobox |
|
PHOX2B
|
ENSG00000109132.5 | paired like homeobox 2B |
|
LHX1
|
ENSG00000132130.7 | LIM homeobox 1 |
|
LMX1A
|
ENSG00000162761.10 | LIM homeobox transcription factor 1 alpha |
|
LHX5
|
ENSG00000089116.3 | LIM homeobox 5 |
|
HOXC4
|
ENSG00000198353.6 | homeobox C4 |
|
HOXC4
|
ENSG00000273266.1 | homeobox C4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC4 | hg19_v2_chr12_+_54410664_54410715 | -0.77 | 7.6e-02 | Click! |
| LHX5 | hg19_v2_chr12_-_113909877_113909877 | -0.65 | 1.7e-01 | Click! |
| LHX1 | hg19_v2_chr17_+_35294075_35294102 | 0.11 | 8.3e-01 | Click! |
Activity profile of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4 motif
Sorted Z-values of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_126409159 | 0.29 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr2_-_43266680 | 0.20 |
ENST00000425212.1
ENST00000422351.1 ENST00000449766.1 |
AC016735.2
|
AC016735.2 |
| chr6_+_130339710 | 0.19 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr2_-_241497390 | 0.16 |
ENST00000272972.3
ENST00000401804.1 ENST00000361678.4 ENST00000405523.3 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chr7_-_150020750 | 0.16 |
ENST00000539352.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr19_-_14064114 | 0.14 |
ENST00000585607.1
ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1
|
podocan-like 1 |
| chr7_-_84122033 | 0.13 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chrX_+_51486481 | 0.13 |
ENST00000340438.4
|
GSPT2
|
G1 to S phase transition 2 |
| chr7_-_150020814 | 0.13 |
ENST00000477871.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr7_+_5465382 | 0.12 |
ENST00000609130.1
|
RP11-1275H24.2
|
RP11-1275H24.2 |
| chr4_-_185275104 | 0.12 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr10_-_4285923 | 0.12 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr2_-_37068530 | 0.12 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr7_-_111032971 | 0.12 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr3_+_186692745 | 0.12 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr10_+_94451574 | 0.11 |
ENST00000492654.2
|
HHEX
|
hematopoietically expressed homeobox |
| chr17_+_1674982 | 0.10 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr12_-_10282836 | 0.10 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr5_+_36606700 | 0.10 |
ENST00000416645.2
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr1_+_225600404 | 0.10 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr5_-_20575959 | 0.10 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr19_+_54466179 | 0.09 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr3_-_141747950 | 0.09 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr4_+_26324474 | 0.09 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr19_-_51920952 | 0.09 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr12_-_54653313 | 0.08 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr16_-_30122717 | 0.08 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr4_+_119810134 | 0.08 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chr17_+_57233087 | 0.08 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr18_-_47018897 | 0.08 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr12_+_4130143 | 0.07 |
ENST00000543206.1
|
RP11-320N7.2
|
RP11-320N7.2 |
| chr16_-_28634874 | 0.07 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr1_+_160370344 | 0.07 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr7_-_44580861 | 0.07 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chr1_+_117544366 | 0.07 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr4_-_36245561 | 0.07 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr19_+_56270380 | 0.06 |
ENST00000434937.2
|
RFPL4A
|
ret finger protein-like 4A |
| chr12_+_4385230 | 0.06 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chr17_+_48823896 | 0.06 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr2_-_241497374 | 0.06 |
ENST00000373318.2
ENST00000406958.1 ENST00000391987.1 ENST00000373320.4 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chr9_-_116065551 | 0.06 |
ENST00000297894.5
|
RNF183
|
ring finger protein 183 |
| chr14_-_51027838 | 0.06 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr4_+_119809984 | 0.06 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr4_+_169418255 | 0.06 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr3_-_149293990 | 0.06 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr11_-_107729504 | 0.06 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr12_+_59989918 | 0.06 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr12_-_25348007 | 0.06 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr13_-_36050819 | 0.05 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr11_-_107729287 | 0.05 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr4_+_86525299 | 0.05 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr20_-_33735070 | 0.05 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr12_+_34175398 | 0.05 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr4_-_177116772 | 0.05 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr18_-_47018869 | 0.05 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr12_+_25205666 | 0.05 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr2_+_166095898 | 0.05 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr10_-_25304889 | 0.05 |
ENST00000483339.2
|
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr3_+_186353756 | 0.05 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr6_-_41039567 | 0.05 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr20_+_57594309 | 0.04 |
ENST00000217133.1
|
TUBB1
|
tubulin, beta 1 class VI |
| chr12_-_64062583 | 0.04 |
ENST00000542209.1
|
DPY19L2
|
dpy-19-like 2 (C. elegans) |
| chr20_+_15177480 | 0.04 |
ENST00000402914.1
|
MACROD2
|
MACRO domain containing 2 |
| chr1_+_62439037 | 0.04 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr11_-_128894053 | 0.04 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chrX_+_55744228 | 0.04 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr16_+_15489603 | 0.04 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr15_+_75970672 | 0.04 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr17_-_8198636 | 0.04 |
ENST00000577745.1
ENST00000579192.1 ENST00000396278.1 |
SLC25A35
|
solute carrier family 25, member 35 |
| chr14_-_104387888 | 0.04 |
ENST00000286953.3
|
C14orf2
|
chromosome 14 open reading frame 2 |
| chr17_+_19091325 | 0.04 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr20_-_50418972 | 0.04 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr7_-_144435985 | 0.04 |
ENST00000549981.1
|
TPK1
|
thiamin pyrophosphokinase 1 |
| chr14_+_50234309 | 0.04 |
ENST00000298307.5
|
KLHDC2
|
kelch domain containing 2 |
| chr17_+_18086392 | 0.04 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr11_-_107729887 | 0.04 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr9_+_125133315 | 0.04 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr11_+_35222629 | 0.04 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr18_-_47018769 | 0.04 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr12_+_104337515 | 0.04 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr9_+_12695702 | 0.04 |
ENST00000381136.2
|
TYRP1
|
tyrosinase-related protein 1 |
| chr12_-_118796971 | 0.04 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr12_+_75784850 | 0.03 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr4_-_74486109 | 0.03 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr16_-_66583701 | 0.03 |
ENST00000527800.1
ENST00000525974.1 ENST00000563369.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr6_+_42584847 | 0.03 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr2_-_136678123 | 0.03 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chrM_-_14670 | 0.03 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr9_-_99540328 | 0.03 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr13_+_36050881 | 0.03 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr1_+_205682497 | 0.03 |
ENST00000598338.1
|
AC119673.1
|
AC119673.1 |
| chr1_+_66820058 | 0.03 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr5_-_41794313 | 0.03 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr15_+_84115868 | 0.03 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr5_+_60933634 | 0.03 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr12_-_112614506 | 0.03 |
ENST00000548588.2
|
HECTD4
|
HECT domain containing E3 ubiquitin protein ligase 4 |
| chr3_-_9994021 | 0.03 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr6_-_109702885 | 0.03 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr15_+_93443419 | 0.03 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr2_-_74618907 | 0.03 |
ENST00000421392.1
ENST00000437375.1 |
DCTN1
|
dynactin 1 |
| chr7_+_30589829 | 0.03 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr14_-_104387831 | 0.03 |
ENST00000557040.1
ENST00000414262.2 ENST00000555030.1 ENST00000554713.1 ENST00000553430.1 |
C14orf2
|
chromosome 14 open reading frame 2 |
| chr15_-_98417780 | 0.03 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr9_-_28670283 | 0.03 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr11_-_559377 | 0.03 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chr1_-_152131703 | 0.03 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr4_+_95128748 | 0.03 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr17_+_48823975 | 0.03 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr1_-_57045228 | 0.03 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr5_+_81575281 | 0.03 |
ENST00000380167.4
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr4_-_87374330 | 0.03 |
ENST00000511328.1
ENST00000503911.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr7_-_22862406 | 0.03 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr19_+_107471 | 0.03 |
ENST00000585993.1
|
OR4F17
|
olfactory receptor, family 4, subfamily F, member 17 |
| chr12_-_112123524 | 0.03 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr10_-_27529486 | 0.03 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr4_+_113568207 | 0.03 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr17_+_67498396 | 0.03 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr15_-_75748115 | 0.03 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr12_-_118796910 | 0.03 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr14_+_55493920 | 0.03 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr4_+_71108300 | 0.03 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr13_+_53602894 | 0.03 |
ENST00000219022.2
|
OLFM4
|
olfactomedin 4 |
| chr4_-_89442940 | 0.02 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr16_+_28565230 | 0.02 |
ENST00000317058.3
|
CCDC101
|
coiled-coil domain containing 101 |
| chr1_+_168250194 | 0.02 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chrX_+_77166172 | 0.02 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr1_-_212965104 | 0.02 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr19_-_36001113 | 0.02 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chrX_-_100872911 | 0.02 |
ENST00000361910.4
ENST00000539247.1 ENST00000538627.1 |
ARMCX6
|
armadillo repeat containing, X-linked 6 |
| chr13_+_103459704 | 0.02 |
ENST00000602836.1
|
BIVM-ERCC5
|
BIVM-ERCC5 readthrough |
| chr5_+_179135246 | 0.02 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr14_+_67831576 | 0.02 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr21_+_17909594 | 0.02 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr6_+_122793058 | 0.02 |
ENST00000392491.2
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chrX_+_55744166 | 0.02 |
ENST00000374941.4
ENST00000414239.1 |
RRAGB
|
Ras-related GTP binding B |
| chr3_-_141719195 | 0.02 |
ENST00000397991.4
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr12_+_16500571 | 0.02 |
ENST00000543076.1
ENST00000396210.3 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr5_+_95066823 | 0.02 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr12_-_15114658 | 0.02 |
ENST00000542276.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr14_+_103851712 | 0.02 |
ENST00000440884.3
ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3
|
MAP/microtubule affinity-regulating kinase 3 |
| chr3_+_139063372 | 0.02 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr12_-_74686314 | 0.02 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr4_+_5527117 | 0.02 |
ENST00000505296.1
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr1_+_78383813 | 0.02 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr14_-_21994337 | 0.02 |
ENST00000537235.1
ENST00000450879.2 |
SALL2
|
spalt-like transcription factor 2 |
| chr1_-_92952433 | 0.02 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr12_+_8309630 | 0.02 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr13_-_81801115 | 0.02 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr8_+_92261516 | 0.02 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chrX_+_43515467 | 0.02 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr14_-_75530693 | 0.02 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr10_+_101542462 | 0.02 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr9_-_26947453 | 0.02 |
ENST00000397292.3
|
PLAA
|
phospholipase A2-activating protein |
| chr18_-_51750948 | 0.02 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr3_-_122233723 | 0.02 |
ENST00000493510.1
ENST00000344337.6 ENST00000476916.1 ENST00000465882.1 |
KPNA1
|
karyopherin alpha 1 (importin alpha 5) |
| chr4_-_39033963 | 0.02 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr10_-_28571015 | 0.02 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr17_-_19015945 | 0.02 |
ENST00000573866.2
|
SNORD3D
|
small nucleolar RNA, C/D box 3D |
| chrX_+_107288239 | 0.02 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chrM_+_14741 | 0.02 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr7_+_129932974 | 0.02 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr20_+_31870927 | 0.02 |
ENST00000253354.1
|
BPIFB1
|
BPI fold containing family B, member 1 |
| chr12_+_16500599 | 0.02 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr21_-_31538971 | 0.02 |
ENST00000286808.3
|
CLDN17
|
claudin 17 |
| chr4_-_76944621 | 0.02 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr4_+_75174180 | 0.02 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr3_-_101039402 | 0.02 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr12_-_10022735 | 0.02 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr6_+_42749759 | 0.02 |
ENST00000314073.5
|
GLTSCR1L
|
GLTSCR1-like |
| chr12_-_15114191 | 0.02 |
ENST00000541380.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr3_-_99569821 | 0.02 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr15_-_37393406 | 0.02 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr2_-_21022818 | 0.02 |
ENST00000440866.2
ENST00000541941.1 ENST00000402479.2 ENST00000435420.2 ENST00000432947.1 ENST00000403006.2 ENST00000419825.2 ENST00000381090.3 ENST00000237822.3 ENST00000412261.1 |
C2orf43
|
chromosome 2 open reading frame 43 |
| chr8_+_29605860 | 0.02 |
ENST00000517491.1
|
AC145110.1
|
AC145110.1 |
| chr9_+_34652164 | 0.02 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr1_-_109935819 | 0.02 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr20_-_50418947 | 0.02 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr7_-_37024665 | 0.02 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr10_-_50396407 | 0.02 |
ENST00000374153.2
ENST00000374151.3 |
C10orf128
|
chromosome 10 open reading frame 128 |
| chr15_+_92006567 | 0.02 |
ENST00000554333.1
|
RP11-661P17.1
|
RP11-661P17.1 |
| chr4_-_120243545 | 0.02 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr21_+_25801041 | 0.01 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chrX_-_106243451 | 0.01 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr15_+_64680003 | 0.01 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr4_+_71091786 | 0.01 |
ENST00000317987.5
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr7_+_73868120 | 0.01 |
ENST00000265755.3
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr4_+_76481258 | 0.01 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr12_+_16500037 | 0.01 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr7_-_22862448 | 0.01 |
ENST00000358435.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr2_-_8977714 | 0.01 |
ENST00000319688.5
ENST00000489024.1 ENST00000256707.3 ENST00000427284.1 ENST00000418530.1 ENST00000473731.1 |
KIDINS220
|
kinase D-interacting substrate, 220kDa |
| chr6_-_27835357 | 0.01 |
ENST00000331442.3
|
HIST1H1B
|
histone cluster 1, H1b |
| chr9_-_44402427 | 0.01 |
ENST00000540551.1
|
BX088651.1
|
LOC100126582 protein; Uncharacterized protein |
| chr9_+_22646189 | 0.01 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr11_+_5710919 | 0.01 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr5_-_88119580 | 0.01 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr10_-_118928543 | 0.01 |
ENST00000419373.2
|
RP11-501J20.2
|
RP11-501J20.2 |
| chr3_+_12329397 | 0.01 |
ENST00000397015.2
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr8_-_62602327 | 0.01 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr4_+_183164574 | 0.01 |
ENST00000511685.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr2_+_30670077 | 0.01 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |