Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
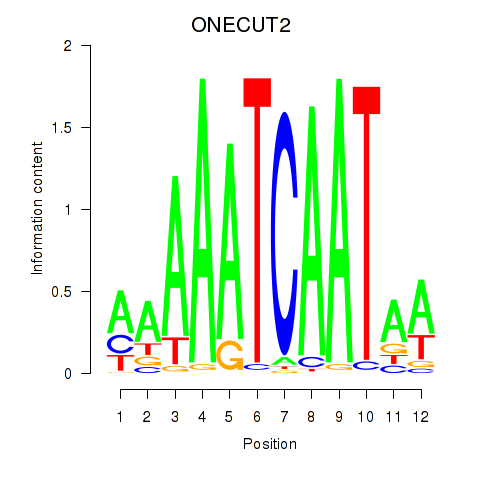
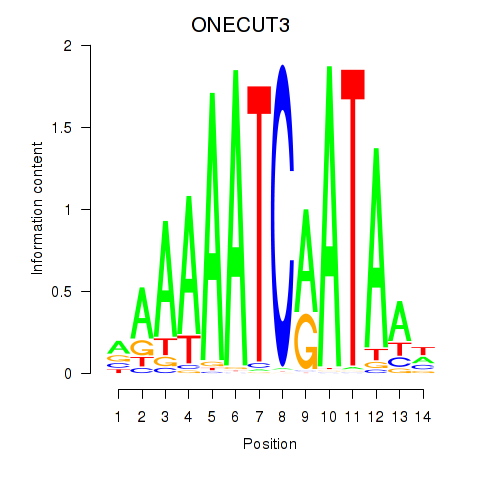
Results for ONECUT2_ONECUT3
Z-value: 0.28
Transcription factors associated with ONECUT2_ONECUT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT2
|
ENSG00000119547.5 | one cut homeobox 2 |
|
ONECUT3
|
ENSG00000205922.4 | one cut homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ONECUT3 | hg19_v2_chr19_+_1752372_1752372 | -0.60 | 2.1e-01 | Click! |
| ONECUT2 | hg19_v2_chr18_+_55102917_55102985 | 0.43 | 3.9e-01 | Click! |
Activity profile of ONECUT2_ONECUT3 motif
Sorted Z-values of ONECUT2_ONECUT3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_61151306 | 0.30 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr12_+_147052 | 0.19 |
ENST00000594563.1
|
AC026369.1
|
Uncharacterized protein |
| chr1_-_116383322 | 0.18 |
ENST00000429731.1
|
NHLH2
|
nescient helix loop helix 2 |
| chr5_+_136070614 | 0.18 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr3_-_186262166 | 0.16 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr14_+_29241910 | 0.16 |
ENST00000399387.4
ENST00000552957.1 ENST00000548213.1 |
C14orf23
|
chromosome 14 open reading frame 23 |
| chr8_+_142264664 | 0.13 |
ENST00000518520.1
|
RP11-10J21.3
|
Uncharacterized protein |
| chrX_-_119693745 | 0.13 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr5_-_33297946 | 0.13 |
ENST00000510327.1
|
CTD-2066L21.3
|
CTD-2066L21.3 |
| chr11_+_47279155 | 0.12 |
ENST00000444396.1
ENST00000457932.1 ENST00000412937.1 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr20_+_58571419 | 0.12 |
ENST00000244049.3
ENST00000350849.6 ENST00000456106.1 |
CDH26
|
cadherin 26 |
| chr7_+_142829162 | 0.11 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr1_+_198608146 | 0.09 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr16_-_4817129 | 0.09 |
ENST00000545009.1
ENST00000219478.6 |
ZNF500
|
zinc finger protein 500 |
| chr5_-_146833803 | 0.08 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr12_+_100594557 | 0.08 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chrX_+_100805496 | 0.08 |
ENST00000372829.3
|
ARMCX1
|
armadillo repeat containing, X-linked 1 |
| chr2_-_206950996 | 0.07 |
ENST00000414320.1
|
INO80D
|
INO80 complex subunit D |
| chr10_+_70980051 | 0.07 |
ENST00000354624.5
ENST00000395086.2 |
HKDC1
|
hexokinase domain containing 1 |
| chr12_+_64846129 | 0.07 |
ENST00000540417.1
ENST00000539810.1 |
TBK1
|
TANK-binding kinase 1 |
| chr1_-_32384693 | 0.07 |
ENST00000602683.1
ENST00000470404.1 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr1_-_116383738 | 0.07 |
ENST00000320238.3
|
NHLH2
|
nescient helix loop helix 2 |
| chr6_+_56954867 | 0.07 |
ENST00000370708.4
ENST00000370702.1 |
ZNF451
|
zinc finger protein 451 |
| chr2_+_67624430 | 0.07 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr12_+_6833237 | 0.07 |
ENST00000229251.3
ENST00000539735.1 ENST00000538410.1 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr12_-_76817036 | 0.06 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr4_-_69536346 | 0.06 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr11_+_63137251 | 0.05 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chr6_+_56954808 | 0.05 |
ENST00000510483.1
ENST00000370706.4 ENST00000357489.3 |
ZNF451
|
zinc finger protein 451 |
| chr11_+_85359002 | 0.05 |
ENST00000528105.1
ENST00000304511.2 |
TMEM126A
|
transmembrane protein 126A |
| chr2_+_105050794 | 0.05 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr9_-_27529726 | 0.05 |
ENST00000262244.5
|
MOB3B
|
MOB kinase activator 3B |
| chr3_-_11685345 | 0.05 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr15_-_76352069 | 0.05 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr12_+_64845864 | 0.05 |
ENST00000538890.1
|
TBK1
|
TANK-binding kinase 1 |
| chr7_+_99425633 | 0.04 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chrX_+_123095860 | 0.04 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr4_-_111563076 | 0.04 |
ENST00000354925.2
ENST00000511990.1 |
PITX2
|
paired-like homeodomain 2 |
| chrX_+_123095890 | 0.04 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr13_+_97928395 | 0.04 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr11_+_85359062 | 0.04 |
ENST00000532180.1
|
TMEM126A
|
transmembrane protein 126A |
| chr3_+_10312604 | 0.04 |
ENST00000426850.1
|
TATDN2
|
TatD DNase domain containing 2 |
| chr17_-_202579 | 0.04 |
ENST00000577079.1
ENST00000331302.7 ENST00000536489.2 |
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr4_-_102267953 | 0.04 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr2_+_58655461 | 0.04 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr2_+_109204909 | 0.04 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr17_+_17685422 | 0.04 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr5_-_143550241 | 0.04 |
ENST00000522203.1
|
YIPF5
|
Yip1 domain family, member 5 |
| chr19_+_35940486 | 0.04 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr1_+_99127225 | 0.04 |
ENST00000370189.5
ENST00000529992.1 |
SNX7
|
sorting nexin 7 |
| chr7_+_133615169 | 0.04 |
ENST00000541309.1
|
EXOC4
|
exocyst complex component 4 |
| chr11_+_28724129 | 0.04 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr6_+_75994709 | 0.03 |
ENST00000438676.1
ENST00000607221.1 |
RP1-234P15.4
|
RP1-234P15.4 |
| chr19_+_7599128 | 0.03 |
ENST00000545201.2
|
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr10_+_69869237 | 0.03 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chrX_-_117119243 | 0.03 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr16_-_20702578 | 0.03 |
ENST00000307493.4
ENST00000219151.4 |
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr20_+_31755934 | 0.03 |
ENST00000354932.5
|
BPIFA2
|
BPI fold containing family A, member 2 |
| chr12_+_86268065 | 0.03 |
ENST00000551529.1
ENST00000256010.6 |
NTS
|
neurotensin |
| chr9_+_140445651 | 0.03 |
ENST00000371443.5
|
MRPL41
|
mitochondrial ribosomal protein L41 |
| chr10_+_71562180 | 0.03 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr5_+_138940742 | 0.03 |
ENST00000398733.3
ENST00000253815.2 ENST00000505007.1 |
UBE2D2
|
ubiquitin-conjugating enzyme E2D 2 |
| chr19_+_31658405 | 0.03 |
ENST00000589511.1
|
CTC-439O9.3
|
CTC-439O9.3 |
| chr4_+_187148556 | 0.03 |
ENST00000264690.6
ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1 |
| chr17_-_79876010 | 0.03 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7 |
| chr7_-_112727774 | 0.03 |
ENST00000297146.3
ENST00000501255.2 |
GPR85
|
G protein-coupled receptor 85 |
| chr3_+_121774202 | 0.03 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr7_-_30008849 | 0.03 |
ENST00000409497.1
|
SCRN1
|
secernin 1 |
| chr2_-_55646412 | 0.03 |
ENST00000413716.2
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr1_-_100598444 | 0.02 |
ENST00000535161.1
ENST00000287482.5 |
SASS6
|
spindle assembly 6 homolog (C. elegans) |
| chrX_-_64196376 | 0.02 |
ENST00000447788.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr7_+_116451100 | 0.02 |
ENST00000464223.1
ENST00000484325.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr10_-_46167722 | 0.02 |
ENST00000374366.3
ENST00000344646.5 |
ZFAND4
|
zinc finger, AN1-type domain 4 |
| chr7_-_23510086 | 0.02 |
ENST00000258729.3
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr6_+_15401075 | 0.02 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr8_-_108510224 | 0.02 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr1_+_207943667 | 0.02 |
ENST00000462968.2
|
CD46
|
CD46 molecule, complement regulatory protein |
| chr10_-_98031310 | 0.02 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr4_-_102268708 | 0.02 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr5_+_128300810 | 0.02 |
ENST00000262462.4
|
SLC27A6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr21_+_39628780 | 0.02 |
ENST00000417042.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr14_+_62164340 | 0.02 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr5_+_147774275 | 0.02 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr17_+_28705921 | 0.02 |
ENST00000225719.4
|
CPD
|
carboxypeptidase D |
| chr7_-_28220354 | 0.02 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr3_+_171561127 | 0.02 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr6_+_56954919 | 0.02 |
ENST00000508603.1
ENST00000491832.2 ENST00000370710.6 |
ZNF451
|
zinc finger protein 451 |
| chr9_-_127710292 | 0.02 |
ENST00000421514.1
|
GOLGA1
|
golgin A1 |
| chr14_-_23426231 | 0.02 |
ENST00000556915.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr6_+_71122974 | 0.02 |
ENST00000418814.2
|
FAM135A
|
family with sequence similarity 135, member A |
| chr21_+_39628655 | 0.02 |
ENST00000398925.1
ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr14_+_37126765 | 0.02 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr7_-_78400598 | 0.02 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr12_-_122018346 | 0.02 |
ENST00000377069.4
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr12_+_81471816 | 0.02 |
ENST00000261206.3
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3 |
| chr5_+_162887556 | 0.02 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr1_-_115259337 | 0.02 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr9_+_95726243 | 0.02 |
ENST00000416701.2
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr5_+_140800638 | 0.02 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr2_-_39348137 | 0.02 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr6_+_114178512 | 0.02 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr15_+_38746307 | 0.02 |
ENST00000397609.2
ENST00000491535.1 |
FAM98B
|
family with sequence similarity 98, member B |
| chr4_-_102268628 | 0.02 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr18_+_42260861 | 0.02 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr4_-_102268484 | 0.02 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr2_+_132479948 | 0.02 |
ENST00000355171.4
|
C2orf27A
|
chromosome 2 open reading frame 27A |
| chr7_-_30009542 | 0.02 |
ENST00000438497.1
|
SCRN1
|
secernin 1 |
| chr14_-_23426322 | 0.02 |
ENST00000555367.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr2_-_220174166 | 0.01 |
ENST00000409251.3
ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr6_-_113754604 | 0.01 |
ENST00000421737.1
|
RP1-124C6.1
|
RP1-124C6.1 |
| chr12_+_32832134 | 0.01 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chr15_-_99789736 | 0.01 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr10_-_98031265 | 0.01 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr3_+_188889737 | 0.01 |
ENST00000345063.3
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr8_+_22601 | 0.01 |
ENST00000522481.3
ENST00000518652.1 |
AC144568.2
|
Uncharacterized protein |
| chr4_-_111563279 | 0.01 |
ENST00000511837.1
|
PITX2
|
paired-like homeodomain 2 |
| chr17_-_46691990 | 0.01 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr19_+_13135386 | 0.01 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr2_-_86850949 | 0.01 |
ENST00000237455.4
|
RNF103
|
ring finger protein 103 |
| chr14_+_37131058 | 0.01 |
ENST00000361487.6
|
PAX9
|
paired box 9 |
| chr10_-_62332357 | 0.01 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr19_+_45596218 | 0.01 |
ENST00000421905.1
ENST00000221462.4 |
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chr1_-_243326612 | 0.01 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr7_-_143059780 | 0.01 |
ENST00000409578.1
ENST00000409346.1 |
FAM131B
|
family with sequence similarity 131, member B |
| chr7_+_129906660 | 0.01 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr2_+_109204743 | 0.01 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr3_+_46283944 | 0.01 |
ENST00000452454.1
ENST00000457243.1 |
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr4_-_69434245 | 0.01 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr14_-_23426337 | 0.01 |
ENST00000342454.8
ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr19_+_9361606 | 0.01 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr19_-_44952635 | 0.01 |
ENST00000592308.1
ENST00000588931.1 ENST00000291187.4 |
ZNF229
|
zinc finger protein 229 |
| chr6_-_79787902 | 0.01 |
ENST00000275034.4
|
PHIP
|
pleckstrin homology domain interacting protein |
| chr9_+_70856397 | 0.01 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr20_-_62587735 | 0.01 |
ENST00000354216.6
ENST00000369892.3 ENST00000358711.3 |
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr8_+_110098850 | 0.01 |
ENST00000518632.1
|
TRHR
|
thyrotropin-releasing hormone receptor |
| chrX_+_46771848 | 0.01 |
ENST00000218343.4
|
PHF16
|
jade family PHD finger 3 |
| chr1_+_153190053 | 0.01 |
ENST00000368744.3
|
PRR9
|
proline rich 9 |
| chr14_+_67831576 | 0.01 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr19_+_37997812 | 0.01 |
ENST00000542455.1
ENST00000587143.1 |
ZNF793
|
zinc finger protein 793 |
| chr19_+_3880581 | 0.01 |
ENST00000450849.2
ENST00000301260.6 ENST00000398448.3 |
ATCAY
|
ataxia, cerebellar, Cayman type |
| chr3_+_130650738 | 0.01 |
ENST00000504612.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr15_-_83837983 | 0.01 |
ENST00000562702.1
|
HDGFRP3
|
Hepatoma-derived growth factor-related protein 3 |
| chr4_-_100212132 | 0.01 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr5_-_143550159 | 0.01 |
ENST00000448443.2
ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5
|
Yip1 domain family, member 5 |
| chr12_+_4699244 | 0.01 |
ENST00000540757.2
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr3_-_141747459 | 0.01 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_-_147610081 | 0.01 |
ENST00000369226.3
|
NBPF24
|
neuroblastoma breakpoint family, member 24 |
| chr1_-_162346657 | 0.01 |
ENST00000367935.5
|
C1orf111
|
chromosome 1 open reading frame 111 |
| chr3_+_69985734 | 0.01 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr16_-_28374829 | 0.01 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr16_+_28763108 | 0.01 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr14_+_69865401 | 0.01 |
ENST00000556605.1
ENST00000336643.5 ENST00000031146.4 |
SLC39A9
|
solute carrier family 39, member 9 |
| chr20_-_22566089 | 0.01 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr3_+_39424828 | 0.01 |
ENST00000273158.4
ENST00000431510.1 |
SLC25A38
|
solute carrier family 25, member 38 |
| chr4_+_156824840 | 0.01 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr1_+_114473350 | 0.01 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr2_-_208030886 | 0.01 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr18_+_11752040 | 0.01 |
ENST00000423027.3
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr14_+_88490894 | 0.01 |
ENST00000556033.1
ENST00000553929.1 ENST00000555996.1 ENST00000556673.1 ENST00000557339.1 ENST00000556684.1 |
RP11-300J18.3
|
long intergenic non-protein coding RNA 1146 |
| chr4_-_103747011 | 0.01 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr17_+_44701402 | 0.01 |
ENST00000575068.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr15_+_86098670 | 0.01 |
ENST00000558811.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr7_-_104909435 | 0.01 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr4_+_169418195 | 0.01 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_-_146068184 | 0.01 |
ENST00000604894.1
ENST00000369323.3 ENST00000479926.2 |
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr7_+_80231466 | 0.01 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr2_-_192016316 | 0.01 |
ENST00000358470.4
ENST00000432798.1 ENST00000450994.1 |
STAT4
|
signal transducer and activator of transcription 4 |
| chr9_+_77230499 | 0.00 |
ENST00000396204.2
|
RORB
|
RAR-related orphan receptor B |
| chr15_-_37392703 | 0.00 |
ENST00000382766.2
ENST00000444725.1 |
MEIS2
|
Meis homeobox 2 |
| chr6_-_151773232 | 0.00 |
ENST00000444024.1
ENST00000367303.4 |
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr12_+_59989791 | 0.00 |
ENST00000552432.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr9_-_139372141 | 0.00 |
ENST00000313050.7
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr2_+_157330081 | 0.00 |
ENST00000409674.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr11_-_60623437 | 0.00 |
ENST00000332539.4
|
PTGDR2
|
prostaglandin D2 receptor 2 |
| chrX_+_123480194 | 0.00 |
ENST00000371139.4
|
SH2D1A
|
SH2 domain containing 1A |
| chr5_-_135701164 | 0.00 |
ENST00000355180.3
ENST00000426057.2 ENST00000513104.1 |
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr16_-_8962200 | 0.00 |
ENST00000562843.1
ENST00000561530.1 ENST00000396593.2 |
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr15_-_37392086 | 0.00 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr6_+_158733692 | 0.00 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr7_-_143059845 | 0.00 |
ENST00000443739.2
|
FAM131B
|
family with sequence similarity 131, member B |
| chr14_-_24020858 | 0.00 |
ENST00000419474.3
|
ZFHX2
|
zinc finger homeobox 2 |
| chrX_-_64196307 | 0.00 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr7_+_129007964 | 0.00 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr6_+_126221034 | 0.00 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr14_-_23426270 | 0.00 |
ENST00000557591.1
ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chrX_-_85302531 | 0.00 |
ENST00000537751.1
ENST00000358786.4 ENST00000357749.2 |
CHM
|
choroideremia (Rab escort protein 1) |
| chr18_+_66465475 | 0.00 |
ENST00000581520.1
|
CCDC102B
|
coiled-coil domain containing 102B |
| chr1_-_46664074 | 0.00 |
ENST00000371986.3
ENST00000371984.3 |
POMGNT1
|
protein O-linked mannose N-acetylglucosaminyltransferase 1 (beta 1,2-) |
| chr12_-_58212487 | 0.00 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr10_-_127505167 | 0.00 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr2_-_32490859 | 0.00 |
ENST00000404025.2
|
NLRC4
|
NLR family, CARD domain containing 4 |
| chr12_+_32832203 | 0.00 |
ENST00000553257.1
ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L
|
dynamin 1-like |
| chr5_-_146833485 | 0.00 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr3_-_64211112 | 0.00 |
ENST00000295902.6
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr22_+_46449674 | 0.00 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr14_+_39736582 | 0.00 |
ENST00000556148.1
ENST00000348007.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr17_+_31318886 | 0.00 |
ENST00000269053.3
ENST00000394638.1 |
SPACA3
|
sperm acrosome associated 3 |
| chr4_-_20985632 | 0.00 |
ENST00000359001.5
|
KCNIP4
|
Kv channel interacting protein 4 |
| chrX_-_19988382 | 0.00 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr2_-_113012592 | 0.00 |
ENST00000272570.5
ENST00000409573.2 |
ZC3H8
|
zinc finger CCCH-type containing 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ONECUT2_ONECUT3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0035701 | immunoglobulin biosynthetic process(GO:0002378) hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.0 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.0 | GO:0018874 | benzoate metabolic process(GO:0018874) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |