Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
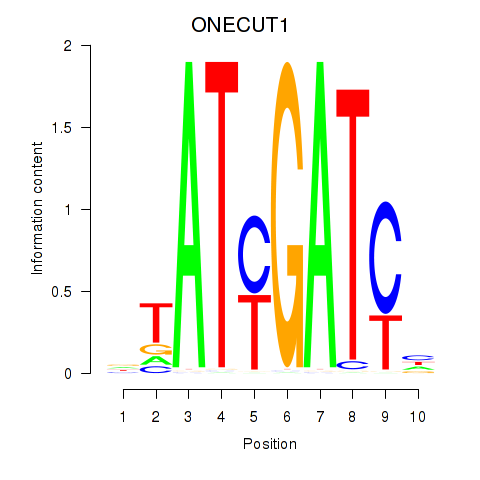
Results for ONECUT1
Z-value: 0.75
Transcription factors associated with ONECUT1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT1
|
ENSG00000169856.7 | one cut homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ONECUT1 | hg19_v2_chr15_-_53082178_53082209 | 0.26 | 6.1e-01 | Click! |
Activity profile of ONECUT1 motif
Sorted Z-values of ONECUT1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_214017151 | 0.62 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr22_+_27068766 | 0.57 |
ENST00000435162.1
ENST00000437071.1 ENST00000440816.1 ENST00000421253.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr16_-_75467274 | 0.52 |
ENST00000566254.1
|
CFDP1
|
craniofacial development protein 1 |
| chr7_+_141463897 | 0.50 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr10_-_53459319 | 0.40 |
ENST00000331173.4
|
CSTF2T
|
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant |
| chr12_-_106477805 | 0.40 |
ENST00000553094.1
ENST00000549704.1 |
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr3_+_32280159 | 0.39 |
ENST00000458535.2
ENST00000307526.3 |
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr10_-_4285923 | 0.33 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr12_+_21679220 | 0.32 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr21_+_42792442 | 0.32 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr3_-_24207039 | 0.30 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr1_-_63782888 | 0.29 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr17_-_26220366 | 0.28 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr22_-_36220420 | 0.27 |
ENST00000473487.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr15_-_86338134 | 0.25 |
ENST00000337975.5
|
KLHL25
|
kelch-like family member 25 |
| chr8_-_23712312 | 0.25 |
ENST00000290271.2
|
STC1
|
stanniocalcin 1 |
| chr3_+_177545563 | 0.23 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr11_+_102188272 | 0.22 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr18_+_47087055 | 0.22 |
ENST00000577628.1
|
LIPG
|
lipase, endothelial |
| chr10_-_4720301 | 0.21 |
ENST00000449712.1
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chrX_-_71792477 | 0.21 |
ENST00000421523.1
ENST00000415409.1 ENST00000373559.4 ENST00000373556.3 ENST00000373560.2 ENST00000373583.1 ENST00000429103.2 ENST00000373571.1 ENST00000373554.1 |
HDAC8
|
histone deacetylase 8 |
| chr1_-_52870104 | 0.21 |
ENST00000371568.3
|
ORC1
|
origin recognition complex, subunit 1 |
| chr15_-_86338100 | 0.21 |
ENST00000536947.1
|
KLHL25
|
kelch-like family member 25 |
| chr2_-_208030295 | 0.21 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr11_-_47788985 | 0.20 |
ENST00000540172.2
|
FNBP4
|
formin binding protein 4 |
| chr1_-_52870059 | 0.20 |
ENST00000371566.1
|
ORC1
|
origin recognition complex, subunit 1 |
| chr18_-_49557 | 0.18 |
ENST00000308911.6
|
RP11-683L23.1
|
Tubulin beta-8 chain-like protein LOC260334 |
| chr1_+_207262881 | 0.18 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr18_+_47087390 | 0.18 |
ENST00000583083.1
|
LIPG
|
lipase, endothelial |
| chr11_+_131240593 | 0.17 |
ENST00000539799.1
|
NTM
|
neurotrimin |
| chr22_+_39101728 | 0.17 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr18_+_39739223 | 0.16 |
ENST00000601948.1
|
LINC00907
|
long intergenic non-protein coding RNA 907 |
| chr6_-_133055815 | 0.16 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chrM_+_14741 | 0.16 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr4_-_83483395 | 0.16 |
ENST00000515780.2
|
TMEM150C
|
transmembrane protein 150C |
| chr5_-_20575959 | 0.16 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr4_-_83483360 | 0.16 |
ENST00000449862.2
|
TMEM150C
|
transmembrane protein 150C |
| chr4_-_103940791 | 0.15 |
ENST00000510559.1
ENST00000394789.3 ENST00000296422.7 |
SLC9B1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr18_+_68002675 | 0.15 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr6_-_31651817 | 0.15 |
ENST00000375863.3
ENST00000375860.2 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr12_-_11422739 | 0.15 |
ENST00000279573.7
|
PRB3
|
proline-rich protein BstNI subfamily 3 |
| chr16_-_75467318 | 0.15 |
ENST00000283882.3
|
CFDP1
|
craniofacial development protein 1 |
| chr21_-_32716556 | 0.14 |
ENST00000455508.1
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1 |
| chr5_-_115910630 | 0.14 |
ENST00000343348.6
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr2_-_200715834 | 0.14 |
ENST00000420128.1
ENST00000416668.1 |
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr12_+_8995832 | 0.14 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr8_+_22601 | 0.14 |
ENST00000522481.3
ENST00000518652.1 |
AC144568.2
|
Uncharacterized protein |
| chr4_+_79475019 | 0.14 |
ENST00000508214.1
|
ANXA3
|
annexin A3 |
| chr14_-_27066960 | 0.14 |
ENST00000539517.2
|
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr6_+_155538093 | 0.13 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr3_+_101546827 | 0.13 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr12_+_70760056 | 0.13 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr1_-_1590418 | 0.13 |
ENST00000341028.7
|
CDK11B
|
cyclin-dependent kinase 11B |
| chrM_-_14670 | 0.13 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr1_+_212738676 | 0.13 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr17_-_26733604 | 0.13 |
ENST00000584426.1
ENST00000584995.1 |
SLC46A1
|
solute carrier family 46 (folate transporter), member 1 |
| chr12_+_69742121 | 0.12 |
ENST00000261267.2
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr16_-_65106110 | 0.12 |
ENST00000562882.1
ENST00000567934.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr1_-_238108575 | 0.12 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr11_+_57365150 | 0.12 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr8_+_95558771 | 0.12 |
ENST00000391679.1
|
AC023632.1
|
HCG2009141; PRO2397; Uncharacterized protein |
| chr3_+_159557637 | 0.12 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr10_+_29135337 | 0.12 |
ENST00000375520.1
|
C10orf126
|
chromosome 10 open reading frame 126 |
| chr11_-_134123142 | 0.12 |
ENST00000392595.2
ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1
|
thymocyte nuclear protein 1 |
| chr1_+_198189921 | 0.12 |
ENST00000391974.3
|
NEK7
|
NIMA-related kinase 7 |
| chr1_-_165414414 | 0.12 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr3_+_127770455 | 0.11 |
ENST00000464451.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr2_-_197457335 | 0.11 |
ENST00000260983.3
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr4_-_17513604 | 0.11 |
ENST00000505710.1
|
QDPR
|
quinoid dihydropteridine reductase |
| chr20_-_58515344 | 0.11 |
ENST00000370996.3
|
PPP1R3D
|
protein phosphatase 1, regulatory subunit 3D |
| chr17_-_26697304 | 0.11 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr10_-_95209 | 0.10 |
ENST00000332708.5
ENST00000309812.4 |
TUBB8
|
tubulin, beta 8 class VIII |
| chr12_+_111471828 | 0.10 |
ENST00000261726.6
|
CUX2
|
cut-like homeobox 2 |
| chr22_-_37505449 | 0.10 |
ENST00000406725.1
|
TMPRSS6
|
transmembrane protease, serine 6 |
| chrX_+_49126294 | 0.10 |
ENST00000466508.1
ENST00000438316.1 ENST00000055335.6 ENST00000495799.1 |
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F |
| chr3_+_32859510 | 0.10 |
ENST00000383763.5
|
TRIM71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr20_+_21686290 | 0.10 |
ENST00000398485.2
|
PAX1
|
paired box 1 |
| chr2_+_234600253 | 0.10 |
ENST00000373424.1
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr9_-_14180778 | 0.10 |
ENST00000380924.1
ENST00000543693.1 |
NFIB
|
nuclear factor I/B |
| chr10_-_28270795 | 0.10 |
ENST00000545014.1
|
ARMC4
|
armadillo repeat containing 4 |
| chr19_-_49243845 | 0.10 |
ENST00000222145.4
|
RASIP1
|
Ras interacting protein 1 |
| chr3_-_112693865 | 0.10 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr10_+_105881779 | 0.10 |
ENST00000369729.3
|
SFR1
|
SWI5-dependent recombination repair 1 |
| chr19_-_13710678 | 0.09 |
ENST00000592864.1
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr16_+_31044413 | 0.09 |
ENST00000394998.1
|
STX4
|
syntaxin 4 |
| chr6_+_126221034 | 0.09 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr1_+_25598872 | 0.09 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
| chr9_+_130830451 | 0.09 |
ENST00000373068.2
ENST00000373069.5 |
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr11_+_134123389 | 0.09 |
ENST00000281182.4
ENST00000537423.1 ENST00000543332.1 ENST00000374752.4 |
ACAD8
|
acyl-CoA dehydrogenase family, member 8 |
| chr12_+_70574088 | 0.09 |
ENST00000552324.1
|
RP11-320P7.2
|
RP11-320P7.2 |
| chr20_+_22034809 | 0.09 |
ENST00000449427.1
|
RP11-125P18.1
|
RP11-125P18.1 |
| chr4_+_169418255 | 0.09 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr4_+_185734773 | 0.09 |
ENST00000508020.1
|
RP11-701P16.2
|
Uncharacterized protein |
| chr12_-_110434021 | 0.09 |
ENST00000355312.3
ENST00000551209.1 ENST00000550186.1 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr3_-_149388682 | 0.09 |
ENST00000475579.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr21_+_39668478 | 0.08 |
ENST00000398927.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr4_+_147145709 | 0.08 |
ENST00000504313.1
|
RP11-6L6.2
|
Uncharacterized protein |
| chr2_-_172967621 | 0.08 |
ENST00000234198.4
ENST00000466293.2 |
DLX2
|
distal-less homeobox 2 |
| chr19_+_11200038 | 0.08 |
ENST00000558518.1
ENST00000557933.1 ENST00000455727.2 ENST00000535915.1 ENST00000545707.1 ENST00000558013.1 |
LDLR
|
low density lipoprotein receptor |
| chr10_+_69869237 | 0.08 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr20_-_22566089 | 0.08 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr1_-_165667545 | 0.08 |
ENST00000538148.1
|
ALDH9A1
|
aldehyde dehydrogenase 9 family, member A1 |
| chr19_-_58864848 | 0.08 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr8_-_33370607 | 0.08 |
ENST00000360742.5
ENST00000523305.1 |
TTI2
|
TELO2 interacting protein 2 |
| chr22_-_37505588 | 0.08 |
ENST00000406856.1
|
TMPRSS6
|
transmembrane protease, serine 6 |
| chrX_+_122318113 | 0.08 |
ENST00000371264.3
|
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr19_+_2785458 | 0.08 |
ENST00000307741.6
ENST00000585338.1 |
THOP1
|
thimet oligopeptidase 1 |
| chr1_-_26633480 | 0.08 |
ENST00000450041.1
|
UBXN11
|
UBX domain protein 11 |
| chr10_-_94003003 | 0.08 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr3_+_98699880 | 0.08 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr7_+_141408153 | 0.08 |
ENST00000397541.2
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr13_-_36920420 | 0.08 |
ENST00000438666.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr12_-_57039739 | 0.08 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chrX_+_122318006 | 0.08 |
ENST00000371266.1
ENST00000264357.5 |
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr16_-_20702578 | 0.07 |
ENST00000307493.4
ENST00000219151.4 |
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr6_-_131299929 | 0.07 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr19_-_48016023 | 0.07 |
ENST00000598615.1
ENST00000597118.1 |
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr17_+_73539232 | 0.07 |
ENST00000580925.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr11_-_122930121 | 0.07 |
ENST00000524552.1
|
HSPA8
|
heat shock 70kDa protein 8 |
| chr17_+_53342311 | 0.07 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chrX_+_10124977 | 0.07 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr6_-_161695074 | 0.07 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr10_-_46167722 | 0.07 |
ENST00000374366.3
ENST00000344646.5 |
ZFAND4
|
zinc finger, AN1-type domain 4 |
| chr10_-_35104185 | 0.07 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr19_+_49588677 | 0.07 |
ENST00000598984.1
ENST00000598441.1 |
SNRNP70
|
small nuclear ribonucleoprotein 70kDa (U1) |
| chr19_+_46800289 | 0.07 |
ENST00000377670.4
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit |
| chr9_-_101471479 | 0.07 |
ENST00000259455.2
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chrX_+_54466829 | 0.07 |
ENST00000375151.4
|
TSR2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr2_+_157330081 | 0.07 |
ENST00000409674.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr9_+_140135665 | 0.07 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr19_+_49588690 | 0.07 |
ENST00000221448.5
|
SNRNP70
|
small nuclear ribonucleoprotein 70kDa (U1) |
| chr16_-_69788816 | 0.07 |
ENST00000268802.5
|
NOB1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr15_-_99789736 | 0.07 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr10_+_63661053 | 0.07 |
ENST00000279873.7
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr19_+_37998031 | 0.07 |
ENST00000586138.1
ENST00000588578.1 ENST00000587986.1 |
ZNF793
|
zinc finger protein 793 |
| chr8_-_108510224 | 0.07 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr19_+_45596398 | 0.07 |
ENST00000544069.2
|
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chr7_+_101459263 | 0.06 |
ENST00000292538.4
ENST00000393824.3 ENST00000547394.2 ENST00000360264.3 ENST00000425244.2 |
CUX1
|
cut-like homeobox 1 |
| chr5_-_75008244 | 0.06 |
ENST00000510798.1
ENST00000446329.2 |
POC5
|
POC5 centriolar protein |
| chr3_+_107364683 | 0.06 |
ENST00000413213.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr14_-_69444957 | 0.06 |
ENST00000556571.1
ENST00000553659.1 ENST00000555616.1 |
ACTN1
|
actinin, alpha 1 |
| chr1_+_25598989 | 0.06 |
ENST00000454452.2
|
RHD
|
Rh blood group, D antigen |
| chr14_-_35873856 | 0.06 |
ENST00000553342.1
ENST00000216797.5 ENST00000557140.1 |
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr15_+_74421961 | 0.06 |
ENST00000565159.1
ENST00000567206.1 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr1_+_152850787 | 0.06 |
ENST00000368765.3
|
SMCP
|
sperm mitochondria-associated cysteine-rich protein |
| chr1_-_25747283 | 0.06 |
ENST00000346452.4
ENST00000340849.4 ENST00000349438.4 ENST00000294413.7 ENST00000413854.1 ENST00000455194.1 ENST00000243186.6 ENST00000425135.1 |
RHCE
|
Rh blood group, CcEe antigens |
| chr5_+_98109322 | 0.06 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr4_-_157892167 | 0.06 |
ENST00000541126.1
|
PDGFC
|
platelet derived growth factor C |
| chr12_-_118490403 | 0.06 |
ENST00000535496.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr5_-_171881491 | 0.06 |
ENST00000311601.5
|
SH3PXD2B
|
SH3 and PX domains 2B |
| chr11_-_72385437 | 0.06 |
ENST00000418754.2
ENST00000542969.2 ENST00000334456.5 |
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr7_+_142985467 | 0.06 |
ENST00000392925.2
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr12_-_96336369 | 0.06 |
ENST00000546947.1
ENST00000546386.1 |
CCDC38
|
coiled-coil domain containing 38 |
| chr12_+_56624436 | 0.06 |
ENST00000266980.4
ENST00000437277.1 |
SLC39A5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr1_-_26633067 | 0.06 |
ENST00000421827.2
ENST00000374215.1 ENST00000374223.1 ENST00000357089.4 ENST00000535108.1 ENST00000314675.7 ENST00000436301.2 ENST00000423664.1 ENST00000374221.3 |
UBXN11
|
UBX domain protein 11 |
| chr5_+_140165876 | 0.06 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr9_-_138853156 | 0.06 |
ENST00000371756.3
|
UBAC1
|
UBA domain containing 1 |
| chr1_-_8075693 | 0.05 |
ENST00000467067.1
|
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr5_+_119799927 | 0.05 |
ENST00000407149.2
ENST00000379551.2 |
PRR16
|
proline rich 16 |
| chr3_+_107364769 | 0.05 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr10_+_97759848 | 0.05 |
ENST00000424464.1
ENST00000410012.2 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr12_+_24857397 | 0.05 |
ENST00000536131.1
|
RP11-625L16.1
|
RP11-625L16.1 |
| chr7_-_154794763 | 0.05 |
ENST00000404141.1
|
PAXIP1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr12_-_13529594 | 0.05 |
ENST00000539026.1
|
C12orf36
|
chromosome 12 open reading frame 36 |
| chr6_-_116866773 | 0.05 |
ENST00000368602.3
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr15_+_48051920 | 0.05 |
ENST00000559196.1
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr7_-_36764142 | 0.05 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr12_+_56623827 | 0.05 |
ENST00000424625.1
ENST00000419753.1 ENST00000454355.2 ENST00000417965.1 ENST00000436633.1 |
SLC39A5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr1_-_1655713 | 0.05 |
ENST00000401096.2
ENST00000404249.3 ENST00000357760.2 ENST00000358779.5 ENST00000378633.1 ENST00000378635.3 |
CDK11A
|
cyclin-dependent kinase 11A |
| chr10_+_90521163 | 0.05 |
ENST00000404459.1
|
LIPN
|
lipase, family member N |
| chr19_+_37997812 | 0.05 |
ENST00000542455.1
ENST00000587143.1 |
ZNF793
|
zinc finger protein 793 |
| chr10_-_45803286 | 0.05 |
ENST00000536058.1
|
OR13A1
|
olfactory receptor, family 13, subfamily A, member 1 |
| chr12_-_54653313 | 0.05 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr11_+_131240373 | 0.05 |
ENST00000374791.3
ENST00000436745.1 |
NTM
|
neurotrimin |
| chrX_+_135388147 | 0.05 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr11_+_107650219 | 0.05 |
ENST00000398067.1
|
AP001024.1
|
Uncharacterized protein |
| chr21_+_17961006 | 0.05 |
ENST00000602323.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr19_-_29218538 | 0.05 |
ENST00000592347.1
ENST00000586528.1 |
AC005307.3
|
AC005307.3 |
| chr12_-_108954933 | 0.05 |
ENST00000431469.2
ENST00000546815.1 |
SART3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr1_+_113933581 | 0.05 |
ENST00000307546.9
ENST00000369615.1 ENST00000369611.4 |
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr1_+_113933371 | 0.05 |
ENST00000369617.4
|
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chrX_-_33229636 | 0.05 |
ENST00000357033.4
|
DMD
|
dystrophin |
| chr4_+_76649753 | 0.05 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr21_-_39870339 | 0.04 |
ENST00000429727.2
ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chrX_+_134654540 | 0.04 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr12_-_95941987 | 0.04 |
ENST00000537435.2
|
USP44
|
ubiquitin specific peptidase 44 |
| chrM_+_8366 | 0.04 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr14_+_62585332 | 0.04 |
ENST00000554895.1
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
| chr15_-_74421477 | 0.04 |
ENST00000514871.1
|
RP11-247C2.2
|
HCG2004779; Uncharacterized protein |
| chr8_-_135522425 | 0.04 |
ENST00000521673.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr12_+_101988627 | 0.04 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr7_+_129007964 | 0.04 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chrM_+_8527 | 0.04 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr16_-_18573396 | 0.04 |
ENST00000543392.1
ENST00000381474.3 ENST00000330537.6 |
NOMO2
|
NODAL modulator 2 |
| chr3_+_181429704 | 0.04 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr2_+_108602971 | 0.04 |
ENST00000409059.1
ENST00000540517.1 ENST00000264047.2 |
SLC5A7
|
solute carrier family 5 (sodium/choline cotransporter), member 7 |
| chr6_+_73076432 | 0.04 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr16_-_30621663 | 0.04 |
ENST00000287461.3
|
ZNF689
|
zinc finger protein 689 |
| chr1_-_151804314 | 0.04 |
ENST00000318247.6
|
RORC
|
RAR-related orphan receptor C |
| chr11_-_122931881 | 0.04 |
ENST00000526110.1
ENST00000227378.3 |
HSPA8
|
heat shock 70kDa protein 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ONECUT1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0010983 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.1 | 0.2 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.3 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.7 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.4 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:1902568 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.4 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.1 | GO:1902871 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.3 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.2 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.0 | 0.1 | GO:0045916 | complement activation, lectin pathway(GO:0001867) negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.0 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.0 | 0.0 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |