Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
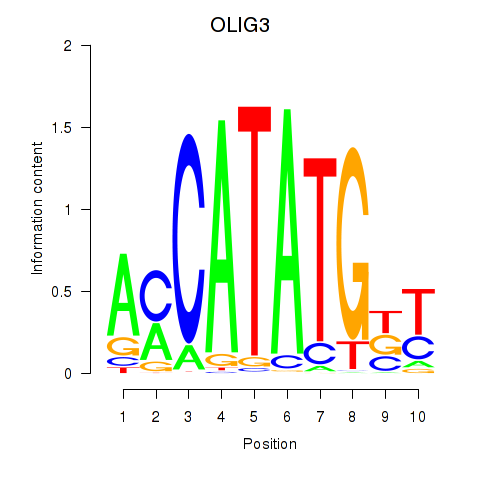
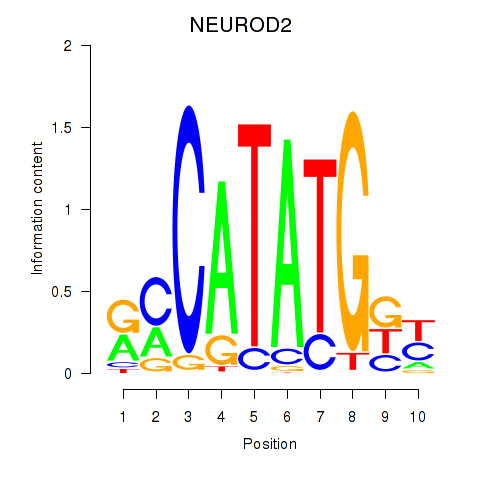
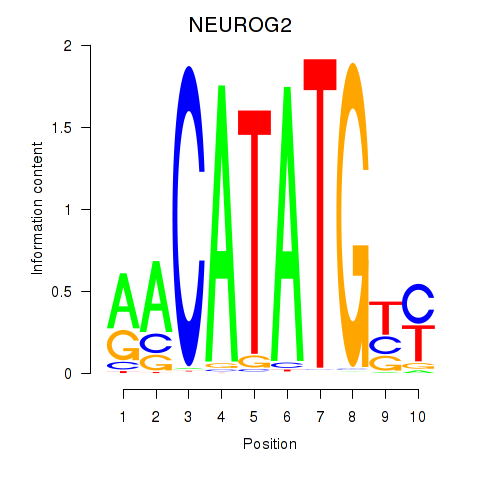
Results for OLIG3_NEUROD2_NEUROG2
Z-value: 0.91
Transcription factors associated with OLIG3_NEUROD2_NEUROG2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG3
|
ENSG00000177468.5 | oligodendrocyte transcription factor 3 |
|
NEUROD2
|
ENSG00000171532.4 | neuronal differentiation 2 |
|
NEUROG2
|
ENSG00000178403.3 | neurogenin 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NEUROD2 | hg19_v2_chr17_-_37764128_37764258 | -0.49 | 3.2e-01 | Click! |
Activity profile of OLIG3_NEUROD2_NEUROG2 motif
Sorted Z-values of OLIG3_NEUROD2_NEUROG2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_66918558 | 0.94 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr19_+_42041860 | 0.82 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr7_-_41742697 | 0.64 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr1_+_95975672 | 0.56 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr15_-_63448973 | 0.55 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr3_+_172361483 | 0.55 |
ENST00000598405.1
|
AC007919.2
|
HCG1787166; PRO1163; Uncharacterized protein |
| chr4_-_140544386 | 0.51 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr12_-_113658892 | 0.45 |
ENST00000299732.2
ENST00000416617.2 |
IQCD
|
IQ motif containing D |
| chr12_+_42624050 | 0.44 |
ENST00000601185.1
|
AC020629.1
|
Uncharacterized protein |
| chr6_-_160147925 | 0.44 |
ENST00000535561.1
|
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr17_-_47925379 | 0.43 |
ENST00000352793.2
ENST00000334568.4 ENST00000398154.1 ENST00000436235.1 ENST00000326219.5 |
TAC4
|
tachykinin 4 (hemokinin) |
| chr1_-_33116128 | 0.42 |
ENST00000436661.1
ENST00000373501.2 ENST00000341885.5 ENST00000468695.1 |
ZBTB8OS
|
zinc finger and BTB domain containing 8 opposite strand |
| chr3_-_197300194 | 0.42 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr15_-_80189380 | 0.41 |
ENST00000258874.3
|
MTHFS
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr16_-_14726576 | 0.39 |
ENST00000562896.1
|
PARN
|
poly(A)-specific ribonuclease |
| chr19_+_57874835 | 0.38 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr19_-_11450249 | 0.38 |
ENST00000222120.3
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr1_+_81106951 | 0.37 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr10_-_69597828 | 0.36 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr6_+_3000057 | 0.36 |
ENST00000397717.2
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr2_+_96068436 | 0.33 |
ENST00000445649.1
ENST00000447036.1 ENST00000233379.4 ENST00000418606.1 |
FAHD2A
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr16_+_2016821 | 0.33 |
ENST00000569210.2
ENST00000569714.1 |
RNF151
|
ring finger protein 151 |
| chrX_+_69509870 | 0.33 |
ENST00000374388.3
|
KIF4A
|
kinesin family member 4A |
| chr12_+_54393880 | 0.31 |
ENST00000303450.4
|
HOXC9
|
homeobox C9 |
| chr12_+_2912363 | 0.30 |
ENST00000544366.1
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr2_-_97760576 | 0.30 |
ENST00000414820.1
ENST00000272610.3 |
FAHD2B
|
fumarylacetoacetate hydrolase domain containing 2B |
| chr2_+_223162866 | 0.29 |
ENST00000295226.1
|
CCDC140
|
coiled-coil domain containing 140 |
| chr18_-_46784778 | 0.29 |
ENST00000582399.1
|
DYM
|
dymeclin |
| chr7_-_24957699 | 0.29 |
ENST00000441059.1
ENST00000415162.1 |
OSBPL3
|
oxysterol binding protein-like 3 |
| chr12_+_133657461 | 0.29 |
ENST00000412146.2
ENST00000544426.1 ENST00000440984.2 ENST00000319849.3 ENST00000440550.2 |
ZNF140
|
zinc finger protein 140 |
| chr14_-_23292596 | 0.28 |
ENST00000554741.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr3_+_42897512 | 0.28 |
ENST00000493193.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr19_-_6057282 | 0.28 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr3_+_185300270 | 0.28 |
ENST00000430355.1
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr6_-_31648150 | 0.27 |
ENST00000375858.3
ENST00000383237.4 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr3_-_131753830 | 0.27 |
ENST00000429747.1
|
CPNE4
|
copine IV |
| chr14_+_91709279 | 0.26 |
ENST00000554096.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr3_-_178984759 | 0.26 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr9_-_10612703 | 0.26 |
ENST00000463477.1
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D |
| chrX_-_46187069 | 0.26 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr10_-_52000307 | 0.25 |
ENST00000443575.1
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr11_-_4629367 | 0.25 |
ENST00000533021.1
|
TRIM68
|
tripartite motif containing 68 |
| chr11_+_18287801 | 0.25 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr12_-_113658826 | 0.24 |
ENST00000546692.1
|
IQCD
|
IQ motif containing D |
| chr3_-_108672742 | 0.24 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr15_-_65503801 | 0.24 |
ENST00000261883.4
|
CILP
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr11_+_18287721 | 0.23 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr14_-_69864993 | 0.23 |
ENST00000555373.1
|
ERH
|
enhancer of rudimentary homolog (Drosophila) |
| chr3_+_113251143 | 0.21 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr11_-_10920838 | 0.21 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr3_+_171758344 | 0.21 |
ENST00000336824.4
ENST00000423424.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr5_+_71475449 | 0.21 |
ENST00000504492.1
|
MAP1B
|
microtubule-associated protein 1B |
| chr2_-_113999260 | 0.21 |
ENST00000468980.2
|
PAX8
|
paired box 8 |
| chr2_+_172543967 | 0.21 |
ENST00000534253.2
ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr9_+_108424738 | 0.19 |
ENST00000334077.3
|
TAL2
|
T-cell acute lymphocytic leukemia 2 |
| chr3_-_121379739 | 0.19 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr5_+_72509751 | 0.19 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr2_+_196440692 | 0.19 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr5_+_67535647 | 0.19 |
ENST00000520675.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr6_+_31887761 | 0.19 |
ENST00000413154.1
|
C2
|
complement component 2 |
| chr16_-_55867146 | 0.19 |
ENST00000422046.2
|
CES1
|
carboxylesterase 1 |
| chr6_+_155443048 | 0.19 |
ENST00000535583.1
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr6_+_135502501 | 0.18 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chrX_+_69509927 | 0.18 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chr1_-_169396666 | 0.18 |
ENST00000456107.1
ENST00000367805.3 |
CCDC181
|
coiled-coil domain containing 181 |
| chr7_-_55930443 | 0.18 |
ENST00000388975.3
|
SEPT14
|
septin 14 |
| chr11_+_120107344 | 0.18 |
ENST00000260264.4
|
POU2F3
|
POU class 2 homeobox 3 |
| chr3_+_38347427 | 0.18 |
ENST00000273173.4
|
SLC22A14
|
solute carrier family 22, member 14 |
| chr14_-_69444957 | 0.18 |
ENST00000556571.1
ENST00000553659.1 ENST00000555616.1 |
ACTN1
|
actinin, alpha 1 |
| chr10_+_44101850 | 0.18 |
ENST00000361807.3
ENST00000374437.2 ENST00000430885.1 ENST00000374435.3 |
ZNF485
|
zinc finger protein 485 |
| chr5_-_150466692 | 0.18 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr6_-_150219232 | 0.18 |
ENST00000531073.1
|
RAET1E
|
retinoic acid early transcript 1E |
| chr17_+_77681075 | 0.18 |
ENST00000397549.2
|
CTD-2116F7.1
|
CTD-2116F7.1 |
| chrX_-_13835398 | 0.18 |
ENST00000475307.1
|
GPM6B
|
glycoprotein M6B |
| chr8_+_142264664 | 0.17 |
ENST00000518520.1
|
RP11-10J21.3
|
Uncharacterized protein |
| chr3_+_98699880 | 0.17 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr1_-_235098861 | 0.17 |
ENST00000458044.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr2_-_214017151 | 0.17 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr6_+_32407619 | 0.17 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr17_+_39846114 | 0.17 |
ENST00000586699.1
|
EIF1
|
eukaryotic translation initiation factor 1 |
| chr8_+_35649365 | 0.16 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr6_+_37400974 | 0.16 |
ENST00000455891.1
ENST00000373451.4 |
CMTR1
|
cap methyltransferase 1 |
| chr18_-_2982869 | 0.16 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr6_+_163148161 | 0.16 |
ENST00000337019.3
ENST00000366889.2 |
PACRG
|
PARK2 co-regulated |
| chr19_+_42041702 | 0.16 |
ENST00000487420.2
|
AC006129.4
|
AC006129.4 |
| chr17_+_46048497 | 0.16 |
ENST00000583352.1
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr2_+_172544011 | 0.16 |
ENST00000508530.1
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr2_+_177053307 | 0.16 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr13_+_49794474 | 0.16 |
ENST00000218721.1
ENST00000398307.1 |
MLNR
|
motilin receptor |
| chr11_+_32112431 | 0.16 |
ENST00000054950.3
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr21_+_42792442 | 0.16 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr1_+_220863187 | 0.16 |
ENST00000294889.5
|
C1orf115
|
chromosome 1 open reading frame 115 |
| chr5_+_157158205 | 0.16 |
ENST00000231198.7
|
THG1L
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr6_+_41010293 | 0.15 |
ENST00000373161.1
ENST00000373158.2 ENST00000470917.1 |
TSPO2
|
translocator protein 2 |
| chr6_-_33714667 | 0.15 |
ENST00000293756.4
|
IP6K3
|
inositol hexakisphosphate kinase 3 |
| chr3_+_187420101 | 0.15 |
ENST00000449623.1
ENST00000437407.1 |
RP11-211G3.3
|
Uncharacterized protein |
| chr1_+_47489240 | 0.15 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr19_+_12035878 | 0.15 |
ENST00000254321.5
ENST00000538752.1 ENST00000590798.1 |
ZNF700
ZNF763
ZNF763
|
zinc finger protein 700 zinc finger protein 763 Uncharacterized protein; Zinc finger protein 763 |
| chr1_+_206808918 | 0.15 |
ENST00000367108.3
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr11_-_96239990 | 0.15 |
ENST00000511243.2
|
JRKL-AS1
|
JRKL antisense RNA 1 |
| chr12_-_91573132 | 0.15 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr21_-_34914394 | 0.15 |
ENST00000361093.5
ENST00000381815.4 |
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr10_+_78078088 | 0.15 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chr4_-_89619386 | 0.15 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr5_-_150460539 | 0.15 |
ENST00000520931.1
ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr1_+_206808868 | 0.15 |
ENST00000367109.2
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr2_+_90458201 | 0.14 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr2_+_219840955 | 0.14 |
ENST00000598002.1
ENST00000432733.1 |
LINC00608
|
long intergenic non-protein coding RNA 608 |
| chr4_+_77941685 | 0.14 |
ENST00000506731.1
|
SEPT11
|
septin 11 |
| chr1_+_145883868 | 0.14 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr2_-_37501692 | 0.14 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr18_-_6928495 | 0.14 |
ENST00000580197.1
|
LINC00668
|
long intergenic non-protein coding RNA 668 |
| chr8_-_37411648 | 0.14 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr5_+_110407390 | 0.14 |
ENST00000344895.3
|
TSLP
|
thymic stromal lymphopoietin |
| chr16_+_14726672 | 0.14 |
ENST00000261658.2
ENST00000563971.1 |
BFAR
|
bifunctional apoptosis regulator |
| chr3_+_137717571 | 0.14 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chr12_-_91546926 | 0.13 |
ENST00000550758.1
|
DCN
|
decorin |
| chr17_+_45331184 | 0.13 |
ENST00000559488.1
ENST00000571680.1 ENST00000435993.2 |
ITGB3
|
integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) |
| chr17_-_37009882 | 0.13 |
ENST00000378096.3
ENST00000394332.1 ENST00000394333.1 ENST00000577407.1 ENST00000479035.2 |
RPL23
|
ribosomal protein L23 |
| chr7_-_107968999 | 0.13 |
ENST00000456431.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr2_+_105953972 | 0.13 |
ENST00000410049.1
|
C2orf49
|
chromosome 2 open reading frame 49 |
| chr12_+_27849378 | 0.13 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr3_-_52719546 | 0.13 |
ENST00000439181.1
ENST00000449505.1 |
PBRM1
|
polybromo 1 |
| chr14_+_32414059 | 0.13 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr5_-_115872124 | 0.13 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr12_-_95941987 | 0.12 |
ENST00000537435.2
|
USP44
|
ubiquitin specific peptidase 44 |
| chr3_+_99833755 | 0.12 |
ENST00000489081.1
|
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr12_+_6898638 | 0.12 |
ENST00000011653.4
|
CD4
|
CD4 molecule |
| chr17_-_58469687 | 0.12 |
ENST00000590133.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr2_+_108994466 | 0.12 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr1_-_82726817 | 0.12 |
ENST00000420549.1
|
RP11-147G16.1
|
RP11-147G16.1 |
| chr2_-_175462934 | 0.12 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr19_+_56905024 | 0.12 |
ENST00000591172.1
ENST00000589888.1 ENST00000587979.1 ENST00000585659.1 ENST00000593109.1 |
ZNF582-AS1
|
ZNF582 antisense RNA 1 (head to head) |
| chr10_-_97416400 | 0.12 |
ENST00000371224.2
ENST00000371221.3 |
ALDH18A1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr12_+_113376249 | 0.12 |
ENST00000551007.1
ENST00000548514.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr10_-_48055018 | 0.12 |
ENST00000426610.2
|
ASAH2C
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2C |
| chr11_-_8795787 | 0.12 |
ENST00000528196.1
ENST00000533681.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr3_-_24207039 | 0.12 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr3_+_185300391 | 0.11 |
ENST00000545472.1
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr1_-_220219775 | 0.11 |
ENST00000609181.1
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr11_+_7595136 | 0.11 |
ENST00000529575.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr1_-_169396646 | 0.11 |
ENST00000367806.3
|
CCDC181
|
coiled-coil domain containing 181 |
| chr12_+_117013656 | 0.11 |
ENST00000556529.1
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr11_-_47870091 | 0.11 |
ENST00000526870.1
|
NUP160
|
nucleoporin 160kDa |
| chr7_-_27205136 | 0.11 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chrX_-_19817869 | 0.11 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr2_+_201994042 | 0.11 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_-_182545603 | 0.11 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr16_-_11922665 | 0.11 |
ENST00000573319.1
ENST00000577041.1 ENST00000574028.1 ENST00000571259.1 ENST00000573037.1 ENST00000571158.1 |
BCAR4
|
breast cancer anti-estrogen resistance 4 (non-protein coding) |
| chr1_-_11118896 | 0.11 |
ENST00000465788.1
|
SRM
|
spermidine synthase |
| chr19_-_36705547 | 0.10 |
ENST00000304116.5
|
ZNF565
|
zinc finger protein 565 |
| chr17_+_65375082 | 0.10 |
ENST00000584471.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr14_-_81425828 | 0.10 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr20_-_35329063 | 0.10 |
ENST00000422536.1
|
NDRG3
|
NDRG family member 3 |
| chr22_-_36556821 | 0.10 |
ENST00000531095.1
ENST00000397293.2 ENST00000349314.2 |
APOL3
|
apolipoprotein L, 3 |
| chr11_+_134146635 | 0.10 |
ENST00000431683.2
|
GLB1L3
|
galactosidase, beta 1-like 3 |
| chr4_+_175205100 | 0.10 |
ENST00000515299.1
|
CEP44
|
centrosomal protein 44kDa |
| chr3_-_52486841 | 0.10 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr2_+_172543919 | 0.10 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr3_+_63638372 | 0.10 |
ENST00000496807.1
|
SNTN
|
sentan, cilia apical structure protein |
| chr4_-_100575781 | 0.10 |
ENST00000511828.1
|
RP11-766F14.2
|
Protein LOC285556 |
| chr15_-_55700216 | 0.10 |
ENST00000569205.1
|
CCPG1
|
cell cycle progression 1 |
| chr11_-_110167331 | 0.10 |
ENST00000534683.1
|
RDX
|
radixin |
| chr6_+_30539153 | 0.10 |
ENST00000326195.8
ENST00000376545.3 ENST00000396515.4 ENST00000441867.1 ENST00000468958.1 |
ABCF1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr3_-_148939835 | 0.10 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr18_-_55253871 | 0.10 |
ENST00000382873.3
|
FECH
|
ferrochelatase |
| chr18_+_61554932 | 0.10 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr1_-_156722195 | 0.10 |
ENST00000368206.5
|
HDGF
|
hepatoma-derived growth factor |
| chr4_+_160188889 | 0.10 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr1_-_21377447 | 0.10 |
ENST00000374937.3
ENST00000264211.8 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr3_-_18480173 | 0.10 |
ENST00000414509.1
|
SATB1
|
SATB homeobox 1 |
| chr11_-_4629388 | 0.10 |
ENST00000526337.1
ENST00000300747.5 |
TRIM68
|
tripartite motif containing 68 |
| chr15_-_83952071 | 0.10 |
ENST00000569704.1
|
BNC1
|
basonuclin 1 |
| chr6_+_46761118 | 0.10 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr8_+_21823726 | 0.10 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr9_-_127177703 | 0.10 |
ENST00000259457.3
ENST00000536392.1 ENST00000441097.1 |
PSMB7
|
proteasome (prosome, macropain) subunit, beta type, 7 |
| chr6_-_28891709 | 0.10 |
ENST00000377194.3
ENST00000377199.3 |
TRIM27
|
tripartite motif containing 27 |
| chr18_-_49557 | 0.10 |
ENST00000308911.6
|
RP11-683L23.1
|
Tubulin beta-8 chain-like protein LOC260334 |
| chr1_+_225600404 | 0.09 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr17_+_66511540 | 0.09 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr18_-_55253830 | 0.09 |
ENST00000591215.1
|
FECH
|
ferrochelatase |
| chrX_-_69509738 | 0.09 |
ENST00000374454.1
ENST00000239666.4 |
PDZD11
|
PDZ domain containing 11 |
| chr19_+_8117881 | 0.09 |
ENST00000390669.3
|
CCL25
|
chemokine (C-C motif) ligand 25 |
| chr8_+_103540983 | 0.09 |
ENST00000523572.1
|
KB-1980E6.3
|
Uncharacterized protein |
| chr7_-_80551671 | 0.09 |
ENST00000419255.2
ENST00000544525.1 |
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr12_+_113416191 | 0.09 |
ENST00000342315.4
ENST00000392583.2 |
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr1_-_120935894 | 0.09 |
ENST00000369383.4
ENST00000369384.4 |
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr8_+_103991013 | 0.09 |
ENST00000517983.1
|
KB-1507C5.4
|
KB-1507C5.4 |
| chr14_+_71679350 | 0.09 |
ENST00000553621.1
|
RP6-91H8.1
|
RP6-91H8.1 |
| chr4_-_74088800 | 0.09 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr1_-_21377383 | 0.09 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr9_-_132404374 | 0.09 |
ENST00000277459.4
ENST00000450050.2 ENST00000277458.4 |
ASB6
|
ankyrin repeat and SOCS box containing 6 |
| chr11_+_134123389 | 0.09 |
ENST00000281182.4
ENST00000537423.1 ENST00000543332.1 ENST00000374752.4 |
ACAD8
|
acyl-CoA dehydrogenase family, member 8 |
| chrX_+_107288197 | 0.09 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr16_-_30905584 | 0.09 |
ENST00000380317.4
|
BCL7C
|
B-cell CLL/lymphoma 7C |
| chr3_-_33700933 | 0.09 |
ENST00000480013.1
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr19_+_35417798 | 0.09 |
ENST00000303586.7
ENST00000439785.1 ENST00000601540.1 |
ZNF30
|
zinc finger protein 30 |
| chr3_-_111314230 | 0.09 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr9_-_14910990 | 0.09 |
ENST00000380881.4
ENST00000422223.2 |
FREM1
|
FRAS1 related extracellular matrix 1 |
| chr14_-_21490653 | 0.09 |
ENST00000449431.2
|
NDRG2
|
NDRG family member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG3_NEUROD2_NEUROG2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.1 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.1 | 0.4 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.4 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.4 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.4 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.2 | GO:1900204 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) thyroid-stimulating hormone secretion(GO:0070460) kidney field specification(GO:0072004) DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.4 | GO:0009397 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.2 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 0.2 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.3 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.2 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 0.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.4 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.2 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.2 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.5 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.2 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.0 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.0 | 0.0 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.6 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0045799 | negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.4 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.2 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) regulation of respiratory burst involved in inflammatory response(GO:0060264) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.1 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.1 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.0 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.0 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.0 | GO:0060262 | regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.0 | 0.1 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.0 | 0.0 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.5 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:2000672 | cellular response to sorbitol(GO:0072709) negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.0 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0051891 | His-Purkinje system development(GO:0003164) cell migration involved in vasculogenesis(GO:0035441) positive regulation of cardioblast differentiation(GO:0051891) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.6 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.0 | GO:0036029 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.6 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.3 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.2 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 0.2 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.1 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.1 | 0.2 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.1 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.4 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0004325 | ferrochelatase activity(GO:0004325) |
| 0.0 | 0.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.1 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.0 | 0.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0052846 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.0 | 0.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.0 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.0 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |