Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
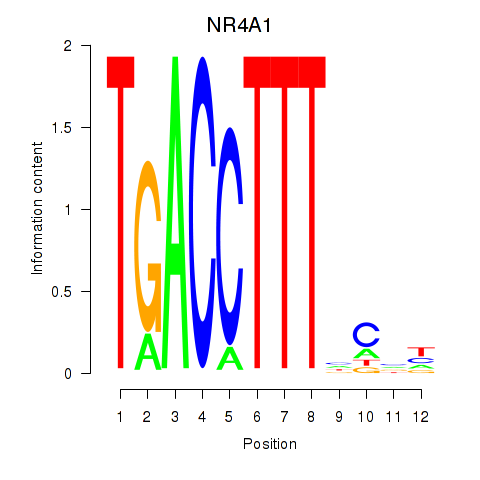
Results for NR4A1
Z-value: 0.74
Transcription factors associated with NR4A1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A1
|
ENSG00000123358.15 | nuclear receptor subfamily 4 group A member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A1 | hg19_v2_chr12_+_52445191_52445243 | -0.59 | 2.1e-01 | Click! |
Activity profile of NR4A1 motif
Sorted Z-values of NR4A1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_141703713 | 0.68 |
ENST00000511815.1
|
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chrX_-_108868390 | 0.59 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr14_+_38065052 | 0.56 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr5_-_126409159 | 0.51 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr17_-_66287310 | 0.44 |
ENST00000582867.1
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr7_-_105516923 | 0.41 |
ENST00000478915.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr6_-_33771757 | 0.39 |
ENST00000507738.1
ENST00000266003.5 ENST00000430124.2 |
MLN
|
motilin |
| chr14_-_104387831 | 0.38 |
ENST00000557040.1
ENST00000414262.2 ENST00000555030.1 ENST00000554713.1 ENST00000553430.1 |
C14orf2
|
chromosome 14 open reading frame 2 |
| chr10_-_33625154 | 0.37 |
ENST00000265371.4
|
NRP1
|
neuropilin 1 |
| chr3_-_113464906 | 0.36 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr2_-_25194476 | 0.35 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr15_+_36887069 | 0.34 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr6_+_30749649 | 0.31 |
ENST00000422944.1
|
HCG20
|
HLA complex group 20 (non-protein coding) |
| chr20_+_12989596 | 0.31 |
ENST00000434210.1
ENST00000399002.2 |
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr2_-_178257401 | 0.30 |
ENST00000464747.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr11_+_86749035 | 0.30 |
ENST00000305494.5
ENST00000535167.1 |
TMEM135
|
transmembrane protein 135 |
| chr11_+_86748863 | 0.28 |
ENST00000340353.7
|
TMEM135
|
transmembrane protein 135 |
| chr15_-_42565023 | 0.28 |
ENST00000566474.1
|
TMEM87A
|
transmembrane protein 87A |
| chr1_+_61869748 | 0.28 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr13_-_33112823 | 0.25 |
ENST00000504114.1
|
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr4_-_40632140 | 0.25 |
ENST00000514782.1
|
RBM47
|
RNA binding motif protein 47 |
| chr14_-_35591433 | 0.23 |
ENST00000261475.5
ENST00000555644.1 |
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr5_-_74162739 | 0.23 |
ENST00000513277.1
|
FAM169A
|
family with sequence similarity 169, member A |
| chr13_-_33112899 | 0.23 |
ENST00000267068.3
ENST00000357505.6 ENST00000399396.3 |
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr19_+_6372444 | 0.22 |
ENST00000245812.3
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr2_+_178257372 | 0.22 |
ENST00000264167.4
ENST00000409888.1 |
AGPS
|
alkylglycerone phosphate synthase |
| chr20_+_12989895 | 0.22 |
ENST00000450297.1
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr16_+_21608525 | 0.20 |
ENST00000567404.1
|
METTL9
|
methyltransferase like 9 |
| chr2_+_162016827 | 0.20 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr11_-_111649074 | 0.20 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr20_+_12989822 | 0.20 |
ENST00000378194.4
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr12_-_95467267 | 0.19 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr14_-_95623607 | 0.19 |
ENST00000531162.1
ENST00000529720.1 ENST00000343455.3 |
DICER1
|
dicer 1, ribonuclease type III |
| chr4_-_40632881 | 0.18 |
ENST00000511598.1
|
RBM47
|
RNA binding motif protein 47 |
| chr13_-_33112956 | 0.18 |
ENST00000505213.1
|
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr11_+_86748957 | 0.18 |
ENST00000526733.1
ENST00000532959.1 |
TMEM135
|
transmembrane protein 135 |
| chr2_+_26568965 | 0.18 |
ENST00000260585.7
ENST00000447170.1 |
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr19_+_17416609 | 0.18 |
ENST00000602206.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr5_-_74162605 | 0.17 |
ENST00000389156.4
ENST00000510496.1 ENST00000380515.3 |
FAM169A
|
family with sequence similarity 169, member A |
| chr6_+_149068464 | 0.16 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr9_-_75567962 | 0.16 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr3_+_29322437 | 0.16 |
ENST00000434693.2
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr5_-_142784003 | 0.16 |
ENST00000416954.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr4_-_111563076 | 0.16 |
ENST00000354925.2
ENST00000511990.1 |
PITX2
|
paired-like homeodomain 2 |
| chr7_-_105517021 | 0.15 |
ENST00000318724.4
ENST00000419735.3 |
ATXN7L1
|
ataxin 7-like 1 |
| chr2_+_162016916 | 0.15 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr3_+_63638339 | 0.15 |
ENST00000343837.3
ENST00000469440.1 |
SNTN
|
sentan, cilia apical structure protein |
| chr16_+_53241854 | 0.15 |
ENST00000565803.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr17_-_79283041 | 0.15 |
ENST00000332012.5
|
LINC00482
|
long intergenic non-protein coding RNA 482 |
| chr15_-_41120896 | 0.15 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr20_+_37590942 | 0.14 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr1_+_64059332 | 0.14 |
ENST00000540265.1
|
PGM1
|
phosphoglucomutase 1 |
| chr14_+_21566980 | 0.14 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr8_-_92053212 | 0.13 |
ENST00000285419.3
|
TMEM55A
|
transmembrane protein 55A |
| chr2_+_234216454 | 0.13 |
ENST00000447536.1
ENST00000409110.1 |
SAG
|
S-antigen; retina and pineal gland (arrestin) |
| chr19_+_17416457 | 0.13 |
ENST00000252602.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr12_+_32655048 | 0.12 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_+_162016804 | 0.12 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr14_-_95624227 | 0.12 |
ENST00000526495.1
|
DICER1
|
dicer 1, ribonuclease type III |
| chr3_-_138763734 | 0.12 |
ENST00000413199.1
ENST00000502927.2 |
PRR23C
|
proline rich 23C |
| chr1_+_212738676 | 0.12 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr3_-_113465065 | 0.12 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chrX_-_43741594 | 0.12 |
ENST00000536181.1
ENST00000378069.4 |
MAOB
|
monoamine oxidase B |
| chr4_-_57253587 | 0.12 |
ENST00000513376.1
ENST00000602986.1 ENST00000434343.2 ENST00000451613.1 ENST00000205214.6 ENST00000502617.1 |
AASDH
|
aminoadipate-semialdehyde dehydrogenase |
| chr7_-_72936531 | 0.12 |
ENST00000339594.4
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr12_+_109577202 | 0.11 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr14_-_35591156 | 0.11 |
ENST00000554361.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr14_-_77495007 | 0.11 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr1_+_176432298 | 0.11 |
ENST00000367661.3
ENST00000367662.3 |
PAPPA2
|
pappalysin 2 |
| chr3_+_113465866 | 0.11 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr12_+_32655110 | 0.10 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr3_-_30936153 | 0.10 |
ENST00000454381.3
ENST00000282538.5 |
GADL1
|
glutamate decarboxylase-like 1 |
| chr4_-_159080806 | 0.10 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr6_-_116381918 | 0.09 |
ENST00000606080.1
|
FRK
|
fyn-related kinase |
| chr4_-_87279641 | 0.09 |
ENST00000512689.1
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr4_-_65275162 | 0.09 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr11_-_111649015 | 0.09 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr2_-_44223138 | 0.09 |
ENST00000260665.7
|
LRPPRC
|
leucine-rich pentatricopeptide repeat containing |
| chr6_+_36973406 | 0.09 |
ENST00000274963.8
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr8_+_133787586 | 0.09 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr7_-_99774945 | 0.09 |
ENST00000292377.2
|
GPC2
|
glypican 2 |
| chr9_+_36572851 | 0.09 |
ENST00000298048.2
ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK
|
maternal embryonic leucine zipper kinase |
| chrX_+_2746850 | 0.08 |
ENST00000381163.3
ENST00000338623.5 ENST00000542787.1 |
GYG2
|
glycogenin 2 |
| chr20_-_42816206 | 0.08 |
ENST00000372980.3
|
JPH2
|
junctophilin 2 |
| chr3_+_14989186 | 0.08 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr2_-_207024134 | 0.07 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr17_+_41150479 | 0.07 |
ENST00000589913.1
|
RPL27
|
ribosomal protein L27 |
| chr4_-_74088800 | 0.07 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr14_+_101295948 | 0.07 |
ENST00000452514.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr14_-_35591679 | 0.07 |
ENST00000557278.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr8_-_92053042 | 0.07 |
ENST00000520014.1
|
TMEM55A
|
transmembrane protein 55A |
| chr1_-_111743285 | 0.06 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr12_-_109219937 | 0.06 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr18_+_43753974 | 0.06 |
ENST00000282059.6
ENST00000321319.6 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr7_-_72936608 | 0.06 |
ENST00000404251.1
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr2_-_25194963 | 0.06 |
ENST00000264711.2
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr6_+_53948221 | 0.06 |
ENST00000460844.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr10_+_101542462 | 0.06 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr4_-_89744314 | 0.06 |
ENST00000508369.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr19_+_4153598 | 0.06 |
ENST00000078445.2
ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3-like 3 |
| chr4_-_40632757 | 0.06 |
ENST00000511902.1
ENST00000505220.1 |
RBM47
|
RNA binding motif protein 47 |
| chr12_-_118490403 | 0.06 |
ENST00000535496.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr4_-_111563279 | 0.06 |
ENST00000511837.1
|
PITX2
|
paired-like homeodomain 2 |
| chr14_+_101295638 | 0.06 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr8_+_123793633 | 0.06 |
ENST00000314393.4
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr4_+_100495864 | 0.05 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr11_-_8680383 | 0.05 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr6_+_30594619 | 0.05 |
ENST00000318999.7
ENST00000376485.4 ENST00000376478.2 ENST00000319027.5 ENST00000376483.4 ENST00000329992.8 ENST00000330083.5 |
ATAT1
|
alpha tubulin acetyltransferase 1 |
| chr6_+_133561725 | 0.05 |
ENST00000452339.2
|
EYA4
|
eyes absent homolog 4 (Drosophila) |
| chr16_+_9185450 | 0.05 |
ENST00000327827.7
|
C16orf72
|
chromosome 16 open reading frame 72 |
| chr3_+_184055240 | 0.05 |
ENST00000383847.2
|
FAM131A
|
family with sequence similarity 131, member A |
| chr7_-_6048702 | 0.05 |
ENST00000265849.7
|
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr2_-_207024233 | 0.05 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr6_+_64346386 | 0.05 |
ENST00000509330.1
|
PHF3
|
PHD finger protein 3 |
| chr4_-_89744365 | 0.05 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr7_-_229557 | 0.05 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr5_-_124084493 | 0.05 |
ENST00000509799.1
|
ZNF608
|
zinc finger protein 608 |
| chr16_+_21312170 | 0.05 |
ENST00000338573.5
ENST00000561968.1 |
CRYM-AS1
|
CRYM antisense RNA 1 |
| chr2_+_207024306 | 0.05 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr14_-_104387888 | 0.05 |
ENST00000286953.3
|
C14orf2
|
chromosome 14 open reading frame 2 |
| chr5_-_87564620 | 0.04 |
ENST00000506536.1
ENST00000512429.1 ENST00000514135.1 ENST00000296595.6 ENST00000509387.1 |
TMEM161B
|
transmembrane protein 161B |
| chr18_-_60986962 | 0.04 |
ENST00000333681.4
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr1_+_165600436 | 0.04 |
ENST00000367888.4
ENST00000367885.1 ENST00000367884.2 |
MGST3
|
microsomal glutathione S-transferase 3 |
| chr15_-_42783303 | 0.04 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr14_+_35591020 | 0.04 |
ENST00000603611.1
|
KIAA0391
|
KIAA0391 |
| chr3_+_101443476 | 0.04 |
ENST00000327230.4
ENST00000494050.1 |
CEP97
|
centrosomal protein 97kDa |
| chr14_+_35591735 | 0.04 |
ENST00000604948.1
ENST00000605201.1 ENST00000250377.7 ENST00000321130.10 ENST00000534898.4 |
KIAA0391
|
KIAA0391 |
| chr3_+_51741072 | 0.04 |
ENST00000395052.3
|
GRM2
|
glutamate receptor, metabotropic 2 |
| chr16_+_691792 | 0.04 |
ENST00000307650.4
|
FAM195A
|
family with sequence similarity 195, member A |
| chr12_+_21679220 | 0.04 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr1_+_104293028 | 0.03 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chrX_+_2746818 | 0.03 |
ENST00000398806.3
|
GYG2
|
glycogenin 2 |
| chr20_+_11898507 | 0.03 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr1_+_174769006 | 0.03 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr4_+_159593271 | 0.03 |
ENST00000512251.1
ENST00000511912.1 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr3_-_44519131 | 0.03 |
ENST00000425708.2
ENST00000396077.2 |
ZNF445
|
zinc finger protein 445 |
| chr5_-_142784101 | 0.03 |
ENST00000503201.1
ENST00000502892.1 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr6_-_152958521 | 0.03 |
ENST00000367255.5
ENST00000265368.4 ENST00000448038.1 ENST00000341594.5 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr18_-_24722995 | 0.03 |
ENST00000581714.1
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr10_-_17171817 | 0.03 |
ENST00000377833.4
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr10_-_61666267 | 0.03 |
ENST00000263102.6
|
CCDC6
|
coiled-coil domain containing 6 |
| chr9_+_12693336 | 0.03 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr11_+_278365 | 0.03 |
ENST00000534750.1
|
NLRP6
|
NLR family, pyrin domain containing 6 |
| chr10_-_103815874 | 0.03 |
ENST00000370033.4
ENST00000311122.5 |
C10orf76
|
chromosome 10 open reading frame 76 |
| chr10_-_75634326 | 0.03 |
ENST00000322635.3
ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr4_-_87279520 | 0.03 |
ENST00000506773.1
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr12_+_32654965 | 0.03 |
ENST00000472289.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr6_-_152489484 | 0.03 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr10_-_75351088 | 0.03 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chrX_-_48823165 | 0.03 |
ENST00000419374.1
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr7_+_120969045 | 0.02 |
ENST00000222462.2
|
WNT16
|
wingless-type MMTV integration site family, member 16 |
| chr17_-_36499693 | 0.02 |
ENST00000342292.4
|
GPR179
|
G protein-coupled receptor 179 |
| chr7_+_6048856 | 0.02 |
ENST00000223029.3
ENST00000400479.2 ENST00000395236.2 |
AIMP2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr3_+_184038073 | 0.02 |
ENST00000428387.1
ENST00000434061.2 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr12_-_118490217 | 0.02 |
ENST00000542304.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr17_+_7123125 | 0.02 |
ENST00000356839.5
ENST00000583312.1 ENST00000350303.5 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr7_-_37488834 | 0.01 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr1_+_16062820 | 0.01 |
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34 |
| chr1_-_161102367 | 0.01 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr6_+_31895254 | 0.01 |
ENST00000299367.5
ENST00000442278.2 |
C2
|
complement component 2 |
| chr1_+_173793777 | 0.01 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr8_+_145065521 | 0.01 |
ENST00000534791.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chrX_+_114874727 | 0.01 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr18_-_52969844 | 0.01 |
ENST00000561831.3
|
TCF4
|
transcription factor 4 |
| chr10_-_75634260 | 0.01 |
ENST00000372765.1
ENST00000351293.3 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr5_+_127039075 | 0.01 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr11_-_113577014 | 0.01 |
ENST00000544634.1
ENST00000539732.1 ENST00000538770.1 ENST00000536856.1 ENST00000544476.1 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chr14_+_35591509 | 0.01 |
ENST00000604073.1
|
KIAA0391
|
KIAA0391 |
| chr19_-_41256207 | 0.00 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr11_+_64073699 | 0.00 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr4_-_40632844 | 0.00 |
ENST00000505414.1
|
RBM47
|
RNA binding motif protein 47 |
| chr5_-_142784888 | 0.00 |
ENST00000514699.1
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr12_+_96252706 | 0.00 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr4_+_57253672 | 0.00 |
ENST00000602927.1
|
RP11-646I6.5
|
RP11-646I6.5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 1.0 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.4 | GO:1902378 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) sensory neuron axon guidance(GO:0097374) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 0.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.2 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.4 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.2 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.2 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.7 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.0 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.0 | 0.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.1 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.7 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.7 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |