Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
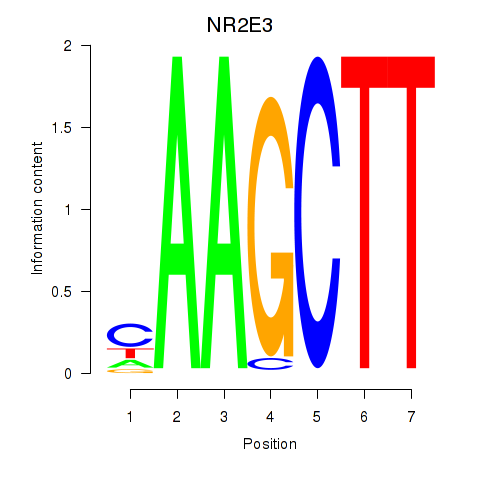
Results for NR2E3
Z-value: 0.92
Transcription factors associated with NR2E3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E3
|
ENSG00000031544.10 | nuclear receptor subfamily 2 group E member 3 |
Activity profile of NR2E3 motif
Sorted Z-values of NR2E3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_148593425 | 0.50 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr14_-_31926623 | 0.50 |
ENST00000356180.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr2_+_145780767 | 0.48 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr11_-_914774 | 0.46 |
ENST00000528154.1
ENST00000525840.1 |
CHID1
|
chitinase domain containing 1 |
| chr3_-_195310802 | 0.40 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr4_-_23891693 | 0.39 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr5_+_59783941 | 0.35 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr5_+_102200948 | 0.35 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chrX_-_106959631 | 0.32 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr4_-_186732892 | 0.32 |
ENST00000451958.1
ENST00000439914.1 ENST00000428330.1 ENST00000429056.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr15_-_41120896 | 0.32 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr3_+_69915363 | 0.31 |
ENST00000451708.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr10_-_91403625 | 0.29 |
ENST00000322191.6
ENST00000342512.3 ENST00000371774.2 |
PANK1
|
pantothenate kinase 1 |
| chr6_+_74405501 | 0.29 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr11_+_121447469 | 0.29 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr3_+_181429704 | 0.28 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr1_+_164528866 | 0.28 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr14_+_54976603 | 0.28 |
ENST00000557317.1
|
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr1_+_149858461 | 0.27 |
ENST00000331380.2
|
HIST2H2AC
|
histone cluster 2, H2ac |
| chr14_+_62462541 | 0.27 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr4_+_174089904 | 0.27 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr12_+_56661461 | 0.27 |
ENST00000546544.1
ENST00000553234.1 |
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr11_-_108408895 | 0.26 |
ENST00000443411.1
ENST00000533052.1 |
EXPH5
|
exophilin 5 |
| chr19_+_2977444 | 0.26 |
ENST00000246112.4
ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr6_+_27215494 | 0.25 |
ENST00000230582.3
|
PRSS16
|
protease, serine, 16 (thymus) |
| chrX_-_117119243 | 0.25 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr6_+_27215471 | 0.25 |
ENST00000421826.2
|
PRSS16
|
protease, serine, 16 (thymus) |
| chr16_+_86612112 | 0.25 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr19_-_40596767 | 0.25 |
ENST00000599972.1
ENST00000450241.2 ENST00000595687.2 |
ZNF780A
|
zinc finger protein 780A |
| chr1_+_164529004 | 0.24 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr12_+_41221975 | 0.24 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chr15_+_42697065 | 0.24 |
ENST00000565559.1
|
CAPN3
|
calpain 3, (p94) |
| chr2_+_190722119 | 0.24 |
ENST00000452382.1
|
PMS1
|
PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
| chr4_+_160188306 | 0.24 |
ENST00000510510.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr15_+_75640068 | 0.24 |
ENST00000565051.1
ENST00000564257.1 ENST00000567005.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr3_+_69915385 | 0.24 |
ENST00000314589.5
|
MITF
|
microphthalmia-associated transcription factor |
| chr2_-_178128250 | 0.23 |
ENST00000448782.1
ENST00000446151.2 |
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr6_-_33679452 | 0.23 |
ENST00000374231.4
ENST00000607484.1 ENST00000374214.3 |
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr5_+_138210919 | 0.23 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr11_-_47600549 | 0.23 |
ENST00000430070.2
|
KBTBD4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr19_-_46706186 | 0.23 |
ENST00000601263.1
ENST00000598754.1 ENST00000594027.1 ENST00000598518.1 ENST00000599817.1 ENST00000595673.1 |
AC006262.5
|
AC006262.5 |
| chr8_+_74903580 | 0.23 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr3_-_196159268 | 0.22 |
ENST00000381887.3
ENST00000535858.1 ENST00000428095.1 ENST00000296328.4 |
UBXN7
|
UBX domain protein 7 |
| chrX_+_123097014 | 0.22 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr1_+_89829610 | 0.22 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chr7_-_124569864 | 0.22 |
ENST00000609702.1
|
POT1
|
protection of telomeres 1 |
| chr20_-_14318248 | 0.22 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr4_+_71859156 | 0.22 |
ENST00000286648.5
ENST00000504730.1 ENST00000504952.1 |
DCK
|
deoxycytidine kinase |
| chr4_+_174089951 | 0.22 |
ENST00000512285.1
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr14_+_31494672 | 0.22 |
ENST00000542754.2
ENST00000313566.6 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr13_+_51913819 | 0.21 |
ENST00000419898.2
|
SERPINE3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr22_-_40929812 | 0.21 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr8_+_67579807 | 0.21 |
ENST00000519289.1
ENST00000519561.1 ENST00000521889.1 |
C8orf44-SGK3
C8orf44
|
C8orf44-SGK3 readthrough chromosome 8 open reading frame 44 |
| chr9_+_72658490 | 0.21 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr6_+_45296391 | 0.21 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr5_-_124084493 | 0.20 |
ENST00000509799.1
|
ZNF608
|
zinc finger protein 608 |
| chr7_-_124569991 | 0.20 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr4_-_88312301 | 0.20 |
ENST00000507286.1
|
HSD17B11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr1_+_84630053 | 0.20 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr11_-_102962874 | 0.20 |
ENST00000531543.1
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 |
| chr20_-_20033052 | 0.20 |
ENST00000536226.1
|
CRNKL1
|
crooked neck pre-mRNA splicing factor 1 |
| chr13_-_86373536 | 0.20 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chrX_-_102565858 | 0.20 |
ENST00000449185.1
ENST00000536889.1 |
BEX2
|
brain expressed X-linked 2 |
| chr5_+_96079240 | 0.20 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr2_+_171036635 | 0.20 |
ENST00000484338.2
ENST00000334231.6 |
MYO3B
|
myosin IIIB |
| chr15_+_45879964 | 0.19 |
ENST00000565409.1
ENST00000564765.1 |
BLOC1S6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr2_-_207082748 | 0.19 |
ENST00000407325.2
ENST00000411719.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr1_-_47131521 | 0.19 |
ENST00000542495.1
ENST00000532925.1 |
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr1_+_62439037 | 0.19 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr18_-_28622699 | 0.19 |
ENST00000360428.4
|
DSC3
|
desmocollin 3 |
| chr13_+_49822041 | 0.19 |
ENST00000538056.1
ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1
|
cytidine and dCMP deaminase domain containing 1 |
| chr5_-_160365601 | 0.18 |
ENST00000518580.1
ENST00000518301.1 |
RP11-109J4.1
|
RP11-109J4.1 |
| chr10_-_98031265 | 0.18 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr2_+_172309634 | 0.18 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr10_-_93392811 | 0.18 |
ENST00000238994.5
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C |
| chr2_-_197675000 | 0.18 |
ENST00000342506.2
|
C2orf66
|
chromosome 2 open reading frame 66 |
| chr11_+_122709200 | 0.18 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr3_-_185826718 | 0.18 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr3_-_165555200 | 0.18 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr19_+_50148087 | 0.18 |
ENST00000601038.1
ENST00000595242.1 |
SCAF1
|
SR-related CTD-associated factor 1 |
| chr1_-_144995074 | 0.18 |
ENST00000534536.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr19_+_17337473 | 0.18 |
ENST00000598068.1
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr12_+_53443963 | 0.18 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr14_-_54908043 | 0.17 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr2_-_207082624 | 0.17 |
ENST00000458440.1
ENST00000437420.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr18_-_28622774 | 0.17 |
ENST00000434452.1
|
DSC3
|
desmocollin 3 |
| chr20_-_40247002 | 0.17 |
ENST00000373222.3
|
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr12_-_15815626 | 0.17 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr3_+_186501979 | 0.17 |
ENST00000498746.1
|
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr4_-_170947565 | 0.17 |
ENST00000506764.1
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr11_+_1891380 | 0.17 |
ENST00000429923.1
ENST00000418975.1 ENST00000406638.2 |
LSP1
|
lymphocyte-specific protein 1 |
| chr7_-_84121858 | 0.17 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr2_-_158732340 | 0.17 |
ENST00000539637.1
ENST00000413751.1 ENST00000434821.1 ENST00000424669.1 |
ACVR1
|
activin A receptor, type I |
| chr12_+_32655110 | 0.17 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr5_+_59783540 | 0.17 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr12_+_41221794 | 0.17 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr9_+_72002837 | 0.16 |
ENST00000377216.3
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr12_+_56661033 | 0.16 |
ENST00000433805.2
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr6_-_113754604 | 0.16 |
ENST00000421737.1
|
RP1-124C6.1
|
RP1-124C6.1 |
| chr1_+_234349957 | 0.16 |
ENST00000366617.3
|
SLC35F3
|
solute carrier family 35, member F3 |
| chr10_-_61122220 | 0.16 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chr8_-_95449155 | 0.16 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr3_-_169381183 | 0.15 |
ENST00000494292.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr4_-_170948361 | 0.15 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr4_-_153332886 | 0.15 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr1_+_47264711 | 0.15 |
ENST00000371923.4
ENST00000271153.4 ENST00000371919.4 |
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1 |
| chr1_+_45140400 | 0.15 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chrX_+_119737806 | 0.15 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chrX_+_105412290 | 0.15 |
ENST00000357175.2
ENST00000337685.2 |
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1 |
| chr5_-_102455801 | 0.15 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr1_-_146697185 | 0.15 |
ENST00000533174.1
ENST00000254090.4 |
FMO5
|
flavin containing monooxygenase 5 |
| chr12_+_28605426 | 0.15 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr19_-_40596828 | 0.15 |
ENST00000414720.2
ENST00000455521.1 ENST00000340963.5 ENST00000595773.1 |
ZNF780A
|
zinc finger protein 780A |
| chr6_-_75953484 | 0.15 |
ENST00000472311.2
ENST00000460985.1 ENST00000377978.3 ENST00000509698.1 ENST00000230459.4 ENST00000370089.2 |
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr2_+_149894968 | 0.15 |
ENST00000409642.3
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr17_+_57233087 | 0.15 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr15_-_33447055 | 0.14 |
ENST00000559047.1
ENST00000561249.1 |
FMN1
|
formin 1 |
| chr3_-_195163584 | 0.14 |
ENST00000439666.1
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr1_+_203595689 | 0.14 |
ENST00000357681.5
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr4_-_129209221 | 0.14 |
ENST00000512483.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr21_+_27543175 | 0.14 |
ENST00000608591.1
ENST00000609365.1 |
AP000230.1
|
AP000230.1 |
| chr6_-_4135693 | 0.14 |
ENST00000495548.1
ENST00000380125.2 ENST00000465828.1 |
ECI2
|
enoyl-CoA delta isomerase 2 |
| chr6_-_4135825 | 0.14 |
ENST00000380118.3
ENST00000413766.2 ENST00000361538.2 |
ECI2
|
enoyl-CoA delta isomerase 2 |
| chr15_-_72565340 | 0.14 |
ENST00000568360.1
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr1_+_156084461 | 0.14 |
ENST00000347559.2
ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA
|
lamin A/C |
| chr17_+_46185111 | 0.14 |
ENST00000582104.1
ENST00000584335.1 |
SNX11
|
sorting nexin 11 |
| chr10_-_103603568 | 0.14 |
ENST00000356640.2
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr12_+_58138664 | 0.13 |
ENST00000257910.3
|
TSPAN31
|
tetraspanin 31 |
| chr9_+_93759317 | 0.13 |
ENST00000563268.1
|
RP11-367F23.2
|
RP11-367F23.2 |
| chr11_-_104905840 | 0.13 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr11_-_47600320 | 0.13 |
ENST00000525720.1
ENST00000531067.1 ENST00000533290.1 ENST00000529499.1 ENST00000529946.1 ENST00000526005.1 ENST00000395288.2 ENST00000534239.1 |
KBTBD4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr2_-_178128149 | 0.13 |
ENST00000423513.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr12_+_32655048 | 0.13 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chrX_-_39956656 | 0.13 |
ENST00000397354.3
ENST00000378444.4 |
BCOR
|
BCL6 corepressor |
| chrX_-_135338503 | 0.13 |
ENST00000370663.5
|
MAP7D3
|
MAP7 domain containing 3 |
| chr5_-_148930731 | 0.13 |
ENST00000515748.2
|
CSNK1A1
|
casein kinase 1, alpha 1 |
| chr11_-_73471655 | 0.13 |
ENST00000400470.2
|
RAB6A
|
RAB6A, member RAS oncogene family |
| chr14_-_61124977 | 0.13 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr12_-_57146095 | 0.13 |
ENST00000550770.1
ENST00000338193.6 |
PRIM1
|
primase, DNA, polypeptide 1 (49kDa) |
| chr1_+_24118452 | 0.13 |
ENST00000421070.1
|
LYPLA2
|
lysophospholipase II |
| chr20_-_40247133 | 0.13 |
ENST00000373233.3
ENST00000309279.7 |
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr12_+_49740700 | 0.12 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr2_-_45795145 | 0.12 |
ENST00000535761.1
|
SRBD1
|
S1 RNA binding domain 1 |
| chr15_+_23255242 | 0.12 |
ENST00000450802.3
|
GOLGA8I
|
golgin A8 family, member I |
| chr18_-_66382289 | 0.12 |
ENST00000443099.2
ENST00000562706.1 ENST00000544714.2 |
TMX3
|
thioredoxin-related transmembrane protein 3 |
| chr11_+_111789580 | 0.12 |
ENST00000278601.5
|
C11orf52
|
chromosome 11 open reading frame 52 |
| chr11_-_82745238 | 0.12 |
ENST00000531021.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr14_-_37641618 | 0.12 |
ENST00000555449.1
|
SLC25A21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr12_+_79258444 | 0.12 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr1_+_84629976 | 0.12 |
ENST00000446538.1
ENST00000370684.1 ENST00000436133.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr15_+_86087267 | 0.12 |
ENST00000558166.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr1_+_168250194 | 0.12 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr2_-_197664366 | 0.12 |
ENST00000409364.3
ENST00000263956.3 |
GTF3C3
|
general transcription factor IIIC, polypeptide 3, 102kDa |
| chr5_+_179135246 | 0.12 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr12_-_25150409 | 0.12 |
ENST00000549262.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr14_+_56127989 | 0.12 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr15_-_38519066 | 0.11 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr4_-_156298087 | 0.11 |
ENST00000311277.4
|
MAP9
|
microtubule-associated protein 9 |
| chrX_+_123094672 | 0.11 |
ENST00000354548.5
ENST00000458700.1 |
STAG2
|
stromal antigen 2 |
| chr10_+_88428370 | 0.11 |
ENST00000372066.3
ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3
|
LIM domain binding 3 |
| chr2_+_160590469 | 0.11 |
ENST00000409591.1
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
| chr1_+_87797351 | 0.11 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr1_+_12042015 | 0.11 |
ENST00000412236.1
|
MFN2
|
mitofusin 2 |
| chr13_+_60971080 | 0.11 |
ENST00000377894.2
|
TDRD3
|
tudor domain containing 3 |
| chr10_-_16563870 | 0.11 |
ENST00000298943.3
|
C1QL3
|
complement component 1, q subcomponent-like 3 |
| chr11_+_17298543 | 0.11 |
ENST00000533926.1
|
NUCB2
|
nucleobindin 2 |
| chr7_+_90339169 | 0.11 |
ENST00000436577.2
|
CDK14
|
cyclin-dependent kinase 14 |
| chr19_-_9546227 | 0.11 |
ENST00000361451.2
ENST00000361151.1 |
ZNF266
|
zinc finger protein 266 |
| chr7_-_103848405 | 0.11 |
ENST00000447452.2
ENST00000545943.1 ENST00000297431.4 |
ORC5
|
origin recognition complex, subunit 5 |
| chr15_-_77197620 | 0.11 |
ENST00000565970.1
ENST00000563290.1 ENST00000565372.1 ENST00000564177.1 ENST00000568382.1 ENST00000563919.1 |
SCAPER
|
S-phase cyclin A-associated protein in the ER |
| chr17_-_57232525 | 0.11 |
ENST00000583380.1
ENST00000580541.1 ENST00000578105.1 ENST00000437036.2 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chrX_-_138914394 | 0.11 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr14_+_74035763 | 0.11 |
ENST00000238651.5
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr3_-_69402828 | 0.10 |
ENST00000460709.1
|
FRMD4B
|
FERM domain containing 4B |
| chr12_+_48516463 | 0.10 |
ENST00000546465.1
|
PFKM
|
phosphofructokinase, muscle |
| chr1_-_229478714 | 0.10 |
ENST00000284617.2
|
CCSAP
|
centriole, cilia and spindle-associated protein |
| chr4_-_25865159 | 0.10 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr5_+_110559312 | 0.10 |
ENST00000508074.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr11_+_74204883 | 0.10 |
ENST00000528481.1
|
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr1_+_84630645 | 0.10 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr15_-_50647347 | 0.10 |
ENST00000220429.8
ENST00000429662.2 |
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr21_-_16374688 | 0.10 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr2_+_130737223 | 0.10 |
ENST00000410061.2
|
RAB6C
|
RAB6C, member RAS oncogene family |
| chr4_+_159727272 | 0.10 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr12_+_32654965 | 0.10 |
ENST00000472289.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr11_+_63997750 | 0.10 |
ENST00000321685.3
|
DNAJC4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr1_-_144995002 | 0.10 |
ENST00000369356.4
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr11_+_17298297 | 0.10 |
ENST00000529010.1
|
NUCB2
|
nucleobindin 2 |
| chr1_+_167298281 | 0.10 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chr17_-_57232596 | 0.10 |
ENST00000581068.1
ENST00000330137.7 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr18_+_23806382 | 0.10 |
ENST00000400466.2
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
| chr19_-_9546177 | 0.10 |
ENST00000592292.1
ENST00000588221.1 |
ZNF266
|
zinc finger protein 266 |
| chr5_-_138211051 | 0.10 |
ENST00000518785.1
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr2_+_73114489 | 0.10 |
ENST00000234454.5
|
SPR
|
sepiapterin reductase (7,8-dihydrobiopterin:NADP+ oxidoreductase) |
| chr6_-_42418999 | 0.10 |
ENST00000340840.2
ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chr7_+_94023873 | 0.10 |
ENST00000297268.6
|
COL1A2
|
collagen, type I, alpha 2 |
| chr12_-_76477707 | 0.09 |
ENST00000551992.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr16_+_21623392 | 0.09 |
ENST00000562961.1
|
METTL9
|
methyltransferase like 9 |
| chr11_+_17298255 | 0.09 |
ENST00000531172.1
ENST00000533738.2 ENST00000323688.6 |
NUCB2
|
nucleobindin 2 |
| chr11_+_17298522 | 0.09 |
ENST00000529313.1
|
NUCB2
|
nucleobindin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.3 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of mitochondrial DNA metabolic process(GO:1901860) response to methionine(GO:1904640) |
| 0.1 | 0.7 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.4 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.3 | GO:1902997 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.4 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.7 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.2 | GO:1903249 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.1 | 0.2 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.1 | 0.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.3 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.6 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.2 | GO:0003274 | endocardial cushion fusion(GO:0003274) |
| 0.0 | 0.3 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.2 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.3 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.2 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 0.0 | 0.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.3 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0030910 | olfactory placode formation(GO:0030910) myotome development(GO:0061055) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.0 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.4 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:1901490 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) regulation of lymphangiogenesis(GO:1901490) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.2 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.0 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.0 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.5 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.0 | GO:1904647 | response to rotenone(GO:1904647) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.4 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.2 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.1 | 0.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.2 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.0 | 0.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.2 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.5 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.9 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.0 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.0 | GO:0004423 | iduronate-2-sulfatase activity(GO:0004423) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |