Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
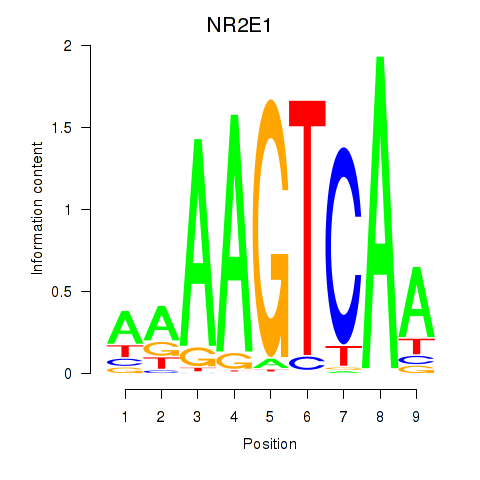
Results for NR2E1
Z-value: 0.61
Transcription factors associated with NR2E1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E1
|
ENSG00000112333.7 | nuclear receptor subfamily 2 group E member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2E1 | hg19_v2_chr6_+_108487245_108487262 | -0.26 | 6.2e-01 | Click! |
Activity profile of NR2E1 motif
Sorted Z-values of NR2E1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_26156551 | 0.31 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr7_+_142829162 | 0.31 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr12_+_72061563 | 0.28 |
ENST00000551238.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr6_-_134861089 | 0.26 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr3_-_138048653 | 0.24 |
ENST00000460099.1
|
NME9
|
NME/NM23 family member 9 |
| chr11_-_62323702 | 0.23 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr12_+_6898674 | 0.23 |
ENST00000541982.1
ENST00000539492.1 |
CD4
|
CD4 molecule |
| chr3_-_43147549 | 0.23 |
ENST00000344697.2
|
POMGNT2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr1_-_48866517 | 0.22 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr3_+_113616317 | 0.22 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr1_-_44818599 | 0.21 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr10_-_21786179 | 0.21 |
ENST00000377113.5
|
CASC10
|
cancer susceptibility candidate 10 |
| chr12_+_122018697 | 0.21 |
ENST00000541574.1
|
RP13-941N14.1
|
RP13-941N14.1 |
| chr14_-_68157084 | 0.20 |
ENST00000557564.1
|
RP11-1012A1.4
|
RP11-1012A1.4 |
| chr4_-_76008706 | 0.20 |
ENST00000562355.1
ENST00000563602.1 |
RP11-44F21.5
|
RP11-44F21.5 |
| chr10_+_85933494 | 0.20 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chrY_+_15418467 | 0.15 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr9_-_75567962 | 0.15 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr3_-_54962100 | 0.15 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr3_-_101232019 | 0.15 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr3_-_138048682 | 0.15 |
ENST00000383180.2
|
NME9
|
NME/NM23 family member 9 |
| chr19_+_39759154 | 0.14 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr6_-_10747802 | 0.14 |
ENST00000606522.1
ENST00000606652.1 |
RP11-421M1.8
|
RP11-421M1.8 |
| chr6_+_142623758 | 0.14 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr17_+_1959369 | 0.14 |
ENST00000576444.1
ENST00000322941.3 |
HIC1
|
hypermethylated in cancer 1 |
| chr12_-_8088871 | 0.14 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr2_+_181988620 | 0.13 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr3_+_121774202 | 0.13 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr6_-_107077347 | 0.13 |
ENST00000369063.3
ENST00000539449.1 |
RTN4IP1
|
reticulon 4 interacting protein 1 |
| chr4_+_183370146 | 0.13 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr5_-_42811986 | 0.13 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr19_+_35940486 | 0.13 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr20_-_22565101 | 0.12 |
ENST00000419308.2
|
FOXA2
|
forkhead box A2 |
| chr9_-_5304432 | 0.12 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr5_-_55777586 | 0.12 |
ENST00000506836.1
|
CTC-236F12.4
|
Uncharacterized protein |
| chrX_+_47444613 | 0.12 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr8_-_37351344 | 0.12 |
ENST00000520422.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr17_+_27895609 | 0.12 |
ENST00000581411.2
ENST00000301057.7 |
TP53I13
|
tumor protein p53 inducible protein 13 |
| chrX_+_10031499 | 0.12 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chrX_+_70364667 | 0.12 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr1_-_152061537 | 0.12 |
ENST00000368806.1
|
TCHHL1
|
trichohyalin-like 1 |
| chr2_-_96926313 | 0.12 |
ENST00000435268.1
|
TMEM127
|
transmembrane protein 127 |
| chr17_+_36886478 | 0.12 |
ENST00000439660.2
|
CISD3
|
CDGSH iron sulfur domain 3 |
| chr14_+_24407940 | 0.11 |
ENST00000354854.1
|
DHRS4-AS1
|
DHRS4-AS1 |
| chr6_+_10723148 | 0.11 |
ENST00000541412.1
ENST00000229563.5 |
TMEM14C
|
transmembrane protein 14C |
| chr9_+_21440440 | 0.11 |
ENST00000276927.1
|
IFNA1
|
interferon, alpha 1 |
| chr16_-_15180257 | 0.11 |
ENST00000540462.1
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr1_-_244006528 | 0.11 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr11_-_111781554 | 0.11 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chr1_-_143767881 | 0.11 |
ENST00000419275.1
|
PPIAL4G
|
peptidylprolyl isomerase A (cyclophilin A)-like 4G |
| chr7_+_134832808 | 0.10 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr16_+_47496023 | 0.10 |
ENST00000567200.1
|
PHKB
|
phosphorylase kinase, beta |
| chr19_+_13134772 | 0.10 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr12_-_68696652 | 0.10 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr16_+_57653989 | 0.10 |
ENST00000567835.1
ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr19_+_13135386 | 0.10 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr9_-_91793675 | 0.10 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr12_-_91572278 | 0.10 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chrX_-_153236620 | 0.10 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr4_+_3388057 | 0.09 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr12_-_8088773 | 0.09 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chrX_+_47077680 | 0.09 |
ENST00000522883.1
|
CDK16
|
cyclin-dependent kinase 16 |
| chr2_-_74875432 | 0.09 |
ENST00000536235.1
ENST00000421985.1 |
M1AP
|
meiosis 1 associated protein |
| chr2_+_67624430 | 0.09 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr20_+_11873141 | 0.09 |
ENST00000422390.1
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr14_-_20774092 | 0.09 |
ENST00000423949.2
ENST00000553828.1 ENST00000258821.3 |
TTC5
|
tetratricopeptide repeat domain 5 |
| chr16_+_3068393 | 0.09 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr14_+_67291158 | 0.09 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr17_-_46667628 | 0.09 |
ENST00000498678.1
|
HOXB3
|
homeobox B3 |
| chr2_-_55237484 | 0.09 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr1_+_114473350 | 0.09 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr1_+_150229554 | 0.09 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr19_-_49567124 | 0.09 |
ENST00000301411.3
|
NTF4
|
neurotrophin 4 |
| chr17_-_46667594 | 0.09 |
ENST00000476342.1
ENST00000460160.1 ENST00000472863.1 |
HOXB3
|
homeobox B3 |
| chr1_+_154975258 | 0.09 |
ENST00000417934.2
|
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chrX_-_138914394 | 0.08 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr13_+_110958124 | 0.08 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr8_-_623547 | 0.08 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chr22_-_33968239 | 0.08 |
ENST00000452586.2
ENST00000421768.1 |
LARGE
|
like-glycosyltransferase |
| chr14_-_71107921 | 0.08 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr13_-_54706954 | 0.08 |
ENST00000606706.1
ENST00000607494.1 ENST00000427299.2 ENST00000423442.2 ENST00000451744.1 |
LINC00458
|
long intergenic non-protein coding RNA 458 |
| chr11_-_111794446 | 0.08 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr10_+_11784360 | 0.08 |
ENST00000379215.4
ENST00000420401.1 |
ECHDC3
|
enoyl CoA hydratase domain containing 3 |
| chr14_+_37131058 | 0.08 |
ENST00000361487.6
|
PAX9
|
paired box 9 |
| chr2_+_101437487 | 0.08 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr8_-_17942432 | 0.08 |
ENST00000381733.4
ENST00000314146.10 |
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| chr1_-_9811600 | 0.08 |
ENST00000435891.1
|
CLSTN1
|
calsyntenin 1 |
| chr20_+_42574317 | 0.08 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chrX_-_131547596 | 0.08 |
ENST00000538204.1
ENST00000370849.3 |
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr20_-_45976816 | 0.08 |
ENST00000441977.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr8_-_124279627 | 0.07 |
ENST00000357082.4
|
ZHX1-C8ORF76
|
ZHX1-C8ORF76 readthrough |
| chr17_+_7533439 | 0.07 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr5_+_173763250 | 0.07 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr19_-_39735646 | 0.07 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr5_-_37371278 | 0.07 |
ENST00000231498.3
|
NUP155
|
nucleoporin 155kDa |
| chr7_+_148936732 | 0.07 |
ENST00000335870.2
|
ZNF212
|
zinc finger protein 212 |
| chr1_-_244013384 | 0.07 |
ENST00000366539.1
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr5_+_177027101 | 0.07 |
ENST00000029410.5
ENST00000510761.1 ENST00000505468.1 |
B4GALT7
|
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 |
| chr2_-_214014959 | 0.07 |
ENST00000442445.1
ENST00000457361.1 ENST00000342002.2 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr4_+_4861385 | 0.07 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr2_-_190446738 | 0.07 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr12_-_122018346 | 0.07 |
ENST00000377069.4
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr4_-_157892055 | 0.07 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr6_+_10747986 | 0.07 |
ENST00000379542.5
|
TMEM14B
|
transmembrane protein 14B |
| chr2_+_131369054 | 0.07 |
ENST00000409602.1
|
POTEJ
|
POTE ankyrin domain family, member J |
| chr3_-_139396787 | 0.07 |
ENST00000296202.7
ENST00000509291.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr13_-_21634421 | 0.07 |
ENST00000542899.1
|
LATS2
|
large tumor suppressor kinase 2 |
| chr17_+_65027509 | 0.07 |
ENST00000375684.1
|
AC005544.1
|
Uncharacterized protein |
| chr6_+_159071015 | 0.07 |
ENST00000360448.3
|
SYTL3
|
synaptotagmin-like 3 |
| chr7_-_105926058 | 0.07 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chrX_-_131547625 | 0.07 |
ENST00000394311.2
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr6_+_10748019 | 0.07 |
ENST00000543878.1
ENST00000461342.1 ENST00000475942.1 ENST00000379530.3 ENST00000473276.1 ENST00000481240.1 ENST00000467317.1 |
SYCP2L
TMEM14B
|
synaptonemal complex protein 2-like transmembrane protein 14B |
| chr5_-_140013224 | 0.06 |
ENST00000498971.2
|
CD14
|
CD14 molecule |
| chr7_+_90012986 | 0.06 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr1_+_161691353 | 0.06 |
ENST00000367948.2
|
FCRLB
|
Fc receptor-like B |
| chr6_+_46661575 | 0.06 |
ENST00000450697.1
|
TDRD6
|
tudor domain containing 6 |
| chr6_+_15246501 | 0.06 |
ENST00000341776.2
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr5_-_87980587 | 0.06 |
ENST00000509783.1
ENST00000509405.1 ENST00000506978.1 ENST00000509265.1 ENST00000513805.1 |
LINC00461
|
long intergenic non-protein coding RNA 461 |
| chr19_-_47220335 | 0.06 |
ENST00000601806.1
ENST00000593363.1 ENST00000598633.1 ENST00000595515.1 ENST00000433867.1 |
PRKD2
|
protein kinase D2 |
| chr7_+_116166331 | 0.06 |
ENST00000393468.1
ENST00000393467.1 |
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr15_+_31196080 | 0.06 |
ENST00000561607.1
ENST00000565466.1 |
FAN1
|
FANCD2/FANCI-associated nuclease 1 |
| chr2_+_11679963 | 0.06 |
ENST00000263834.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr17_+_46133307 | 0.06 |
ENST00000580037.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr4_-_47983519 | 0.06 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr4_+_126315091 | 0.06 |
ENST00000335110.5
|
FAT4
|
FAT atypical cadherin 4 |
| chr5_+_56471592 | 0.06 |
ENST00000511209.1
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr15_-_42783303 | 0.06 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr12_-_89746264 | 0.06 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr6_+_31515337 | 0.06 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chrX_-_106362013 | 0.06 |
ENST00000372487.1
ENST00000372479.3 ENST00000203616.8 |
RBM41
|
RNA binding motif protein 41 |
| chr12_+_40618873 | 0.06 |
ENST00000298910.7
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr10_-_46167722 | 0.05 |
ENST00000374366.3
ENST00000344646.5 |
ZFAND4
|
zinc finger, AN1-type domain 4 |
| chr17_+_19186292 | 0.05 |
ENST00000395626.1
ENST00000571254.1 |
EPN2
|
epsin 2 |
| chr17_+_67498396 | 0.05 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr8_-_116504448 | 0.05 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr9_-_98189055 | 0.05 |
ENST00000433644.2
|
RP11-435O5.2
|
RP11-435O5.2 |
| chr10_+_17794251 | 0.05 |
ENST00000377495.1
ENST00000338221.5 |
TMEM236
|
transmembrane protein 236 |
| chr12_-_111180644 | 0.05 |
ENST00000551676.1
ENST00000550991.1 ENST00000335007.5 ENST00000340766.5 |
PPP1CC
|
protein phosphatase 1, catalytic subunit, gamma isozyme |
| chr3_-_18466787 | 0.05 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr17_-_29624343 | 0.05 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr12_-_120241187 | 0.05 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr2_+_109237717 | 0.05 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr2_+_27440229 | 0.05 |
ENST00000264705.4
ENST00000403525.1 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr4_-_175041663 | 0.05 |
ENST00000503140.1
|
RP11-148L24.1
|
RP11-148L24.1 |
| chr3_-_18466026 | 0.05 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr4_-_153303658 | 0.05 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr20_-_30310656 | 0.05 |
ENST00000376055.4
|
BCL2L1
|
BCL2-like 1 |
| chr17_-_42327236 | 0.05 |
ENST00000399246.2
|
AC003102.1
|
AC003102.1 |
| chr20_-_30310693 | 0.05 |
ENST00000307677.4
ENST00000420653.1 |
BCL2L1
|
BCL2-like 1 |
| chr7_+_150076406 | 0.05 |
ENST00000329630.5
|
ZNF775
|
zinc finger protein 775 |
| chr6_+_125540951 | 0.05 |
ENST00000524679.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr5_-_142780280 | 0.05 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr15_-_34502278 | 0.05 |
ENST00000559515.1
ENST00000256544.3 ENST00000560108.1 ENST00000559462.1 |
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr17_-_40833858 | 0.05 |
ENST00000332438.4
|
CCR10
|
chemokine (C-C motif) receptor 10 |
| chr3_-_33686925 | 0.05 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr22_-_30695471 | 0.04 |
ENST00000434291.1
|
RP1-130H16.18
|
Uncharacterized protein |
| chrX_-_73061339 | 0.04 |
ENST00000602863.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr9_-_128246769 | 0.04 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr6_-_42690312 | 0.04 |
ENST00000230381.5
|
PRPH2
|
peripherin 2 (retinal degeneration, slow) |
| chr2_-_106013400 | 0.04 |
ENST00000409807.1
|
FHL2
|
four and a half LIM domains 2 |
| chr9_-_99064429 | 0.04 |
ENST00000375263.3
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr14_+_31494841 | 0.04 |
ENST00000556232.1
ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr8_+_9953061 | 0.04 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr17_-_73258425 | 0.04 |
ENST00000578348.1
ENST00000582486.1 ENST00000582717.1 |
GGA3
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr2_-_39187479 | 0.04 |
ENST00000601251.1
|
AC019171.1
|
Uncharacterized protein |
| chr3_+_179322481 | 0.04 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr12_+_14518598 | 0.04 |
ENST00000261168.4
ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr4_-_46996424 | 0.04 |
ENST00000264318.3
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr3_-_148939598 | 0.04 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr15_+_58702742 | 0.04 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr7_-_151217166 | 0.04 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr12_-_71031185 | 0.04 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr11_-_83984231 | 0.04 |
ENST00000330014.6
ENST00000537455.1 ENST00000376106.3 ENST00000418306.2 ENST00000531015.1 |
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr1_+_165864800 | 0.04 |
ENST00000469256.2
|
UCK2
|
uridine-cytidine kinase 2 |
| chr12_+_10366223 | 0.04 |
ENST00000545290.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr11_-_111781454 | 0.04 |
ENST00000533280.1
|
CRYAB
|
crystallin, alpha B |
| chr10_-_62060232 | 0.04 |
ENST00000503925.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr12_-_50643664 | 0.04 |
ENST00000550592.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr5_+_147774275 | 0.04 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr12_-_52887034 | 0.04 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr11_+_65851443 | 0.04 |
ENST00000533756.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr6_-_88411911 | 0.04 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr16_+_81272287 | 0.04 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr2_+_207630081 | 0.04 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr17_-_8198636 | 0.04 |
ENST00000577745.1
ENST00000579192.1 ENST00000396278.1 |
SLC25A35
|
solute carrier family 25, member 35 |
| chr12_-_90049828 | 0.04 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr16_+_22019404 | 0.04 |
ENST00000542527.2
ENST00000569656.1 ENST00000562695.1 |
C16orf52
|
chromosome 16 open reading frame 52 |
| chr11_-_63439174 | 0.04 |
ENST00000332645.4
|
ATL3
|
atlastin GTPase 3 |
| chr17_+_29664830 | 0.04 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr7_+_100026406 | 0.04 |
ENST00000414441.1
|
MEPCE
|
methylphosphate capping enzyme |
| chr7_-_151217001 | 0.04 |
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain |
| chr16_+_30087288 | 0.03 |
ENST00000279387.7
ENST00000562664.1 ENST00000562222.1 |
PPP4C
|
protein phosphatase 4, catalytic subunit |
| chr7_+_80267973 | 0.03 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr19_-_46234119 | 0.03 |
ENST00000317683.3
|
FBXO46
|
F-box protein 46 |
| chr5_+_67588391 | 0.03 |
ENST00000523872.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr11_-_66104237 | 0.03 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr18_+_29027696 | 0.03 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr9_-_5830768 | 0.03 |
ENST00000381506.3
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chrX_-_47863348 | 0.03 |
ENST00000376943.3
ENST00000396965.1 ENST00000305127.6 |
ZNF182
|
zinc finger protein 182 |
| chr18_+_77867177 | 0.03 |
ENST00000560752.1
|
ADNP2
|
ADNP homeobox 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0045013 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0035397 | helper T cell enhancement of adaptive immune response(GO:0035397) |
| 0.0 | 0.1 | GO:0008052 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.0 | 0.2 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0070839 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) divalent metal ion export(GO:0070839) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0071727 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.0 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.2 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.3 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |