Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
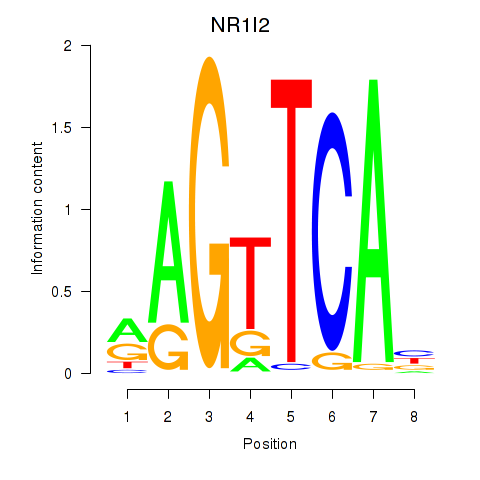
Results for NR1I2
Z-value: 0.94
Transcription factors associated with NR1I2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I2
|
ENSG00000144852.12 | nuclear receptor subfamily 1 group I member 2 |
Activity profile of NR1I2 motif
Sorted Z-values of NR1I2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_67563250 | 0.52 |
ENST00000566907.1
|
FAM65A
|
family with sequence similarity 65, member A |
| chr22_+_22676808 | 0.50 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr3_-_69590486 | 0.45 |
ENST00000497880.1
|
FRMD4B
|
FERM domain containing 4B |
| chr12_+_34175398 | 0.44 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr2_+_181988620 | 0.43 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr6_+_12007963 | 0.40 |
ENST00000607445.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr19_+_2977444 | 0.37 |
ENST00000246112.4
ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr13_-_95364389 | 0.34 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr4_+_183065793 | 0.34 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr13_+_25670268 | 0.33 |
ENST00000281589.3
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3 |
| chr22_+_46546406 | 0.31 |
ENST00000440343.1
ENST00000415785.1 |
PPARA
|
peroxisome proliferator-activated receptor alpha |
| chr7_-_95225768 | 0.29 |
ENST00000005178.5
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr14_+_23016437 | 0.29 |
ENST00000478163.3
|
TRAC
|
T cell receptor alpha constant |
| chr17_+_75137460 | 0.28 |
ENST00000587820.1
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr1_+_16083123 | 0.27 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr9_-_34381536 | 0.26 |
ENST00000379126.3
ENST00000379127.1 ENST00000379133.3 |
C9orf24
|
chromosome 9 open reading frame 24 |
| chr19_-_3547305 | 0.25 |
ENST00000589063.1
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr6_-_152623231 | 0.25 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr5_+_139055021 | 0.25 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr11_-_46141338 | 0.25 |
ENST00000529782.1
ENST00000532010.1 ENST00000525438.1 ENST00000533757.1 ENST00000527782.1 |
PHF21A
|
PHD finger protein 21A |
| chr1_+_24646263 | 0.25 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr19_-_38878632 | 0.25 |
ENST00000586599.1
ENST00000334928.6 ENST00000587676.1 |
GGN
|
gametogenetin |
| chr2_-_235405168 | 0.24 |
ENST00000339728.3
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr18_-_53253112 | 0.23 |
ENST00000568673.1
ENST00000562847.1 ENST00000568147.1 |
TCF4
|
transcription factor 4 |
| chr8_-_101733794 | 0.23 |
ENST00000523555.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr8_-_131028782 | 0.22 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr3_+_133524459 | 0.22 |
ENST00000484684.1
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr2_+_171034646 | 0.21 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr14_-_23395623 | 0.21 |
ENST00000556043.1
|
PRMT5
|
protein arginine methyltransferase 5 |
| chr20_+_62492566 | 0.20 |
ENST00000369916.3
|
ABHD16B
|
abhydrolase domain containing 16B |
| chr14_+_62462541 | 0.20 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr17_+_75123947 | 0.20 |
ENST00000586429.1
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr10_+_90484301 | 0.20 |
ENST00000404190.1
|
LIPK
|
lipase, family member K |
| chr14_-_58893876 | 0.20 |
ENST00000555097.1
ENST00000555404.1 |
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr6_+_142623758 | 0.20 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr2_-_86790472 | 0.20 |
ENST00000409727.1
|
CHMP3
|
charged multivesicular body protein 3 |
| chrX_+_135279179 | 0.19 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr22_+_30821732 | 0.19 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr18_-_53253323 | 0.19 |
ENST00000540999.1
ENST00000563888.2 |
TCF4
|
transcription factor 4 |
| chr12_-_68696652 | 0.19 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr21_-_31588338 | 0.19 |
ENST00000286809.1
|
CLDN8
|
claudin 8 |
| chr2_-_192711968 | 0.19 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr12_-_95397442 | 0.18 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr2_-_86790593 | 0.18 |
ENST00000263856.4
ENST00000409225.2 |
CHMP3
|
charged multivesicular body protein 3 |
| chr14_-_76447494 | 0.18 |
ENST00000238682.3
|
TGFB3
|
transforming growth factor, beta 3 |
| chr1_+_228870824 | 0.18 |
ENST00000366691.3
|
RHOU
|
ras homolog family member U |
| chr1_+_149858461 | 0.18 |
ENST00000331380.2
|
HIST2H2AC
|
histone cluster 2, H2ac |
| chr18_-_52989525 | 0.18 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr8_+_67579807 | 0.18 |
ENST00000519289.1
ENST00000519561.1 ENST00000521889.1 |
C8orf44-SGK3
C8orf44
|
C8orf44-SGK3 readthrough chromosome 8 open reading frame 44 |
| chr1_+_164528437 | 0.18 |
ENST00000485769.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr7_+_97910962 | 0.18 |
ENST00000539286.1
|
BRI3
|
brain protein I3 |
| chrX_+_123097014 | 0.18 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr9_-_96215822 | 0.17 |
ENST00000375412.5
|
FAM120AOS
|
family with sequence similarity 120A opposite strand |
| chr17_-_39123144 | 0.17 |
ENST00000355612.2
|
KRT39
|
keratin 39 |
| chr3_+_112930387 | 0.17 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr5_+_139055055 | 0.17 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chr1_+_16083098 | 0.17 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr15_+_96875657 | 0.17 |
ENST00000559679.1
ENST00000394171.2 |
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr9_+_21440440 | 0.17 |
ENST00000276927.1
|
IFNA1
|
interferon, alpha 1 |
| chr7_-_83278322 | 0.17 |
ENST00000307792.3
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr12_-_6665200 | 0.17 |
ENST00000336604.4
ENST00000396840.2 ENST00000356896.4 |
IFFO1
|
intermediate filament family orphan 1 |
| chr6_+_149068464 | 0.17 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr11_+_86502085 | 0.17 |
ENST00000527521.1
|
PRSS23
|
protease, serine, 23 |
| chr6_+_144980954 | 0.16 |
ENST00000367525.3
|
UTRN
|
utrophin |
| chr4_+_95972822 | 0.16 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr3_-_100566492 | 0.16 |
ENST00000528490.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr8_-_121824374 | 0.16 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr11_-_46639436 | 0.16 |
ENST00000532281.1
|
HARBI1
|
harbinger transposase derived 1 |
| chr3_+_13978886 | 0.15 |
ENST00000524375.1
ENST00000326972.8 |
TPRXL
|
tetra-peptide repeat homeobox-like |
| chr9_-_118417 | 0.15 |
ENST00000382500.2
|
FOXD4
|
forkhead box D4 |
| chr12_-_8088871 | 0.15 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr1_-_55089191 | 0.15 |
ENST00000302250.2
ENST00000371304.2 |
FAM151A
|
family with sequence similarity 151, member A |
| chr2_+_97454321 | 0.15 |
ENST00000540067.1
|
CNNM4
|
cyclin M4 |
| chr4_-_159094194 | 0.15 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr15_+_36887069 | 0.15 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr3_-_49722523 | 0.15 |
ENST00000448220.1
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr11_+_107461948 | 0.15 |
ENST00000265840.7
ENST00000443271.2 |
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr18_-_53255379 | 0.15 |
ENST00000565908.2
|
TCF4
|
transcription factor 4 |
| chr1_-_144995074 | 0.15 |
ENST00000534536.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chrX_+_9880590 | 0.14 |
ENST00000452575.1
|
SHROOM2
|
shroom family member 2 |
| chr17_-_46035187 | 0.14 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr4_+_113970772 | 0.14 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr16_+_83986827 | 0.14 |
ENST00000393306.1
ENST00000565123.1 |
OSGIN1
|
oxidative stress induced growth inhibitor 1 |
| chr7_+_45613958 | 0.14 |
ENST00000297323.7
|
ADCY1
|
adenylate cyclase 1 (brain) |
| chr1_+_61869748 | 0.14 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr18_+_21572737 | 0.14 |
ENST00000304621.6
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr6_+_79577189 | 0.14 |
ENST00000369940.2
|
IRAK1BP1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr3_-_58196939 | 0.14 |
ENST00000394549.2
ENST00000461914.3 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr3_-_113465065 | 0.14 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr4_+_30721968 | 0.14 |
ENST00000361762.2
|
PCDH7
|
protocadherin 7 |
| chr7_+_37960163 | 0.14 |
ENST00000199448.4
ENST00000559325.1 ENST00000423717.1 |
EPDR1
|
ependymin related 1 |
| chrX_+_9880412 | 0.14 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr3_+_37284824 | 0.13 |
ENST00000431105.1
|
GOLGA4
|
golgin A4 |
| chr1_-_153521597 | 0.13 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr12_-_15104040 | 0.13 |
ENST00000541644.1
ENST00000545895.1 |
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr4_+_106067943 | 0.13 |
ENST00000380013.4
ENST00000394764.1 ENST00000413648.2 |
TET2
|
tet methylcytosine dioxygenase 2 |
| chr10_-_76995675 | 0.13 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr9_+_35829208 | 0.13 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr15_+_42697065 | 0.13 |
ENST00000565559.1
|
CAPN3
|
calpain 3, (p94) |
| chr8_-_131028869 | 0.13 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr11_+_122709200 | 0.13 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr7_-_6066183 | 0.13 |
ENST00000422786.1
|
EIF2AK1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr1_+_12538594 | 0.13 |
ENST00000543710.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr3_+_25469802 | 0.13 |
ENST00000330688.4
|
RARB
|
retinoic acid receptor, beta |
| chr10_-_33625154 | 0.13 |
ENST00000265371.4
|
NRP1
|
neuropilin 1 |
| chr1_+_109419804 | 0.13 |
ENST00000435475.1
|
GPSM2
|
G-protein signaling modulator 2 |
| chr17_+_4802713 | 0.13 |
ENST00000521575.1
ENST00000381365.3 |
C17orf107
|
chromosome 17 open reading frame 107 |
| chr17_+_33448593 | 0.13 |
ENST00000158009.5
|
FNDC8
|
fibronectin type III domain containing 8 |
| chr5_-_133747589 | 0.12 |
ENST00000458198.2
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr9_-_14313893 | 0.12 |
ENST00000380921.3
ENST00000380959.3 |
NFIB
|
nuclear factor I/B |
| chr10_-_25010795 | 0.12 |
ENST00000416305.1
ENST00000376410.2 |
ARHGAP21
|
Rho GTPase activating protein 21 |
| chrX_-_48056199 | 0.12 |
ENST00000311798.1
ENST00000347757.1 |
SSX5
|
synovial sarcoma, X breakpoint 5 |
| chr12_-_8088773 | 0.12 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr6_-_27782548 | 0.12 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr2_+_109223595 | 0.12 |
ENST00000410093.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr11_-_108369101 | 0.12 |
ENST00000323468.5
|
KDELC2
|
KDEL (Lys-Asp-Glu-Leu) containing 2 |
| chr17_-_73761222 | 0.12 |
ENST00000437911.1
ENST00000225614.2 |
GALK1
|
galactokinase 1 |
| chr18_-_53253000 | 0.12 |
ENST00000566514.1
|
TCF4
|
transcription factor 4 |
| chr22_+_21128167 | 0.12 |
ENST00000215727.5
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr2_+_219433588 | 0.12 |
ENST00000295701.5
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr5_+_149569520 | 0.12 |
ENST00000230671.2
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr1_+_45274154 | 0.12 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr19_+_7011509 | 0.12 |
ENST00000377296.3
|
AC025278.1
|
Uncharacterized protein |
| chr2_+_17997763 | 0.12 |
ENST00000281047.3
|
MSGN1
|
mesogenin 1 |
| chr6_-_107077347 | 0.12 |
ENST00000369063.3
ENST00000539449.1 |
RTN4IP1
|
reticulon 4 interacting protein 1 |
| chr4_+_71859156 | 0.11 |
ENST00000286648.5
ENST00000504730.1 ENST00000504952.1 |
DCK
|
deoxycytidine kinase |
| chr16_-_58328923 | 0.11 |
ENST00000567164.1
ENST00000219301.4 ENST00000569727.1 |
PRSS54
|
protease, serine, 54 |
| chr3_-_45957088 | 0.11 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr14_+_51026844 | 0.11 |
ENST00000554886.1
|
ATL1
|
atlastin GTPase 1 |
| chr4_+_71600063 | 0.11 |
ENST00000513597.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr4_+_20702030 | 0.11 |
ENST00000510051.1
ENST00000503585.1 ENST00000360916.5 ENST00000295290.8 ENST00000514485.1 |
PACRGL
|
PARK2 co-regulated-like |
| chr4_+_26322409 | 0.11 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr18_-_53255766 | 0.11 |
ENST00000566286.1
ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4
|
transcription factor 4 |
| chr1_-_144994909 | 0.11 |
ENST00000369347.4
ENST00000369354.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr8_+_21911054 | 0.11 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr3_+_25469724 | 0.11 |
ENST00000437042.2
|
RARB
|
retinoic acid receptor, beta |
| chr15_-_75017711 | 0.11 |
ENST00000567032.1
ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr1_-_169555779 | 0.11 |
ENST00000367797.3
ENST00000367796.3 |
F5
|
coagulation factor V (proaccelerin, labile factor) |
| chr6_-_134861089 | 0.11 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr3_-_113464906 | 0.11 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr4_+_20702059 | 0.11 |
ENST00000444671.2
ENST00000510700.1 ENST00000506745.1 ENST00000514663.1 ENST00000509469.1 ENST00000515339.1 ENST00000513861.1 ENST00000502374.1 ENST00000538990.1 ENST00000511160.1 ENST00000504630.1 ENST00000513590.1 ENST00000514292.1 ENST00000502938.1 ENST00000509625.1 ENST00000505160.1 ENST00000507634.1 ENST00000513459.1 ENST00000511089.1 |
PACRGL
|
PARK2 co-regulated-like |
| chr2_+_179149636 | 0.11 |
ENST00000409631.1
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr8_-_18666360 | 0.11 |
ENST00000286485.8
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr15_+_43809797 | 0.11 |
ENST00000399453.1
ENST00000300231.5 |
MAP1A
|
microtubule-associated protein 1A |
| chr18_-_5296001 | 0.11 |
ENST00000357006.4
|
ZBTB14
|
zinc finger and BTB domain containing 14 |
| chr6_-_8102714 | 0.11 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr3_+_137906353 | 0.10 |
ENST00000461822.1
ENST00000485396.1 ENST00000471453.1 ENST00000470821.1 ENST00000471709.1 ENST00000538260.1 ENST00000393058.3 ENST00000463485.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr13_+_97928395 | 0.10 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr15_+_96876340 | 0.10 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr1_+_174769006 | 0.10 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr2_+_111490161 | 0.10 |
ENST00000340561.4
|
ACOXL
|
acyl-CoA oxidase-like |
| chr10_-_120938303 | 0.10 |
ENST00000356951.3
ENST00000298510.2 |
PRDX3
|
peroxiredoxin 3 |
| chr5_+_137203541 | 0.10 |
ENST00000421631.2
|
MYOT
|
myotilin |
| chr2_-_158732340 | 0.10 |
ENST00000539637.1
ENST00000413751.1 ENST00000434821.1 ENST00000424669.1 |
ACVR1
|
activin A receptor, type I |
| chr6_+_106988986 | 0.10 |
ENST00000457437.1
ENST00000535438.1 |
AIM1
|
absent in melanoma 1 |
| chr1_+_40862501 | 0.10 |
ENST00000539317.1
|
SMAP2
|
small ArfGAP2 |
| chr16_-_4164027 | 0.10 |
ENST00000572288.1
|
ADCY9
|
adenylate cyclase 9 |
| chr19_-_46285646 | 0.10 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr3_-_45957534 | 0.10 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr7_-_99381884 | 0.10 |
ENST00000336411.2
|
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chrX_+_102840408 | 0.10 |
ENST00000468024.1
ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4
|
transcription elongation factor A (SII)-like 4 |
| chr8_+_110552831 | 0.09 |
ENST00000530629.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr15_+_59903975 | 0.09 |
ENST00000560585.1
ENST00000396065.1 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr6_+_142622991 | 0.09 |
ENST00000230173.6
ENST00000367608.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr10_-_105677427 | 0.09 |
ENST00000369764.1
|
OBFC1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr2_+_30455016 | 0.09 |
ENST00000401506.1
ENST00000407930.2 |
LBH
|
limb bud and heart development |
| chr15_+_57540213 | 0.09 |
ENST00000559710.1
|
TCF12
|
transcription factor 12 |
| chr2_-_69870747 | 0.09 |
ENST00000409068.1
|
AAK1
|
AP2 associated kinase 1 |
| chr7_-_150777874 | 0.09 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr3_+_69985734 | 0.09 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr12_-_120884175 | 0.09 |
ENST00000546954.1
|
TRIAP1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr4_-_185729602 | 0.09 |
ENST00000437665.3
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr12_-_94673956 | 0.09 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr12_-_6715808 | 0.09 |
ENST00000545584.1
|
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr3_+_153839149 | 0.09 |
ENST00000465093.1
ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr17_-_7082861 | 0.09 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr11_-_63993690 | 0.09 |
ENST00000394546.2
ENST00000541278.1 |
TRPT1
|
tRNA phosphotransferase 1 |
| chr2_+_242127924 | 0.09 |
ENST00000402530.3
ENST00000274979.8 ENST00000402430.3 |
ANO7
|
anoctamin 7 |
| chr6_-_56716686 | 0.09 |
ENST00000520645.1
|
DST
|
dystonin |
| chr8_+_102504651 | 0.09 |
ENST00000251808.3
ENST00000521085.1 |
GRHL2
|
grainyhead-like 2 (Drosophila) |
| chr11_+_120894781 | 0.08 |
ENST00000529397.1
ENST00000528512.1 ENST00000422003.2 |
TBCEL
|
tubulin folding cofactor E-like |
| chr13_-_31191642 | 0.08 |
ENST00000405805.1
|
HMGB1
|
high mobility group box 1 |
| chr5_+_134074191 | 0.08 |
ENST00000297156.2
|
CAMLG
|
calcium modulating ligand |
| chr19_+_42817527 | 0.08 |
ENST00000598766.1
|
TMEM145
|
transmembrane protein 145 |
| chrX_+_28605516 | 0.08 |
ENST00000378993.1
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1 |
| chr13_+_115000339 | 0.08 |
ENST00000360383.3
ENST00000375312.3 |
CDC16
|
cell division cycle 16 |
| chr1_-_205744574 | 0.08 |
ENST00000367139.3
ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr15_+_92397051 | 0.08 |
ENST00000424469.2
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chr5_-_158757895 | 0.08 |
ENST00000231228.2
|
IL12B
|
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
| chr20_-_52790512 | 0.08 |
ENST00000216862.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr6_+_142623063 | 0.08 |
ENST00000296932.8
ENST00000367609.3 |
GPR126
|
G protein-coupled receptor 126 |
| chr20_+_43343886 | 0.08 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr1_-_53387352 | 0.08 |
ENST00000541281.1
|
ECHDC2
|
enoyl CoA hydratase domain containing 2 |
| chr13_-_33112823 | 0.08 |
ENST00000504114.1
|
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr13_-_33112899 | 0.08 |
ENST00000267068.3
ENST00000357505.6 ENST00000399396.3 |
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr9_+_17135016 | 0.08 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chr2_+_166430619 | 0.08 |
ENST00000409420.1
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr3_-_9994021 | 0.08 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr2_+_219433281 | 0.08 |
ENST00000273064.6
ENST00000509807.2 ENST00000542068.1 |
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr11_+_9482551 | 0.08 |
ENST00000438144.2
ENST00000526657.1 ENST00000299606.2 ENST00000534265.1 ENST00000412390.2 |
ZNF143
|
zinc finger protein 143 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1I2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.3 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.1 | 0.4 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.3 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.1 | 0.2 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.3 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.4 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.5 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.2 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.0 | 0.1 | GO:0097374 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) sensory neuron axon guidance(GO:0097374) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.2 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) |
| 0.0 | 0.4 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.1 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.0 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.1 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.3 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0072069 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) thyroid-stimulating hormone secretion(GO:0070460) kidney field specification(GO:0072004) DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.0 | 0.0 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.2 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.0 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.3 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.2 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 0.5 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.3 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.4 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.3 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 1.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.0 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.0 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |