Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
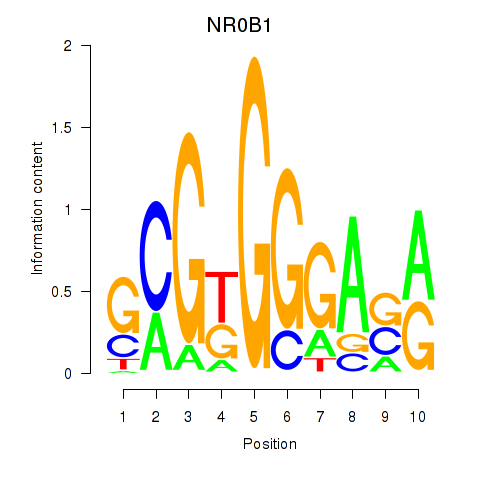
Results for NR0B1
Z-value: 0.91
Transcription factors associated with NR0B1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR0B1
|
ENSG00000169297.6 | nuclear receptor subfamily 0 group B member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR0B1 | hg19_v2_chrX_-_30326445_30326605 | 0.91 | 1.3e-02 | Click! |
Activity profile of NR0B1 motif
Sorted Z-values of NR0B1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_125486939 | 0.71 |
ENST00000303545.3
|
RNF139
|
ring finger protein 139 |
| chr16_+_53164956 | 0.59 |
ENST00000563410.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr1_+_6052700 | 0.55 |
ENST00000378092.1
ENST00000445501.1 |
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr11_-_73471655 | 0.55 |
ENST00000400470.2
|
RAB6A
|
RAB6A, member RAS oncogene family |
| chr3_-_169381166 | 0.53 |
ENST00000486748.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr13_-_76111945 | 0.53 |
ENST00000355801.4
ENST00000406936.3 |
COMMD6
|
COMM domain containing 6 |
| chr1_-_171711177 | 0.49 |
ENST00000415773.1
ENST00000367740.2 |
VAMP4
|
vesicle-associated membrane protein 4 |
| chr7_+_149411860 | 0.48 |
ENST00000486744.1
|
KRBA1
|
KRAB-A domain containing 1 |
| chr19_-_10628098 | 0.48 |
ENST00000590601.1
|
S1PR5
|
sphingosine-1-phosphate receptor 5 |
| chr12_+_56661461 | 0.47 |
ENST00000546544.1
ENST00000553234.1 |
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr8_+_42396712 | 0.41 |
ENST00000518574.1
ENST00000417410.2 ENST00000414154.2 |
SMIM19
|
small integral membrane protein 19 |
| chr3_-_45957088 | 0.37 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr3_-_113464906 | 0.37 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr1_+_212458834 | 0.37 |
ENST00000261461.2
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr2_-_152684977 | 0.36 |
ENST00000428992.2
ENST00000295087.8 |
ARL5A
|
ADP-ribosylation factor-like 5A |
| chr2_-_165698322 | 0.35 |
ENST00000444537.1
ENST00000414843.1 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr17_+_29421987 | 0.34 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr8_+_42396936 | 0.34 |
ENST00000416469.2
|
SMIM19
|
small integral membrane protein 19 |
| chr1_+_44401479 | 0.31 |
ENST00000438616.3
|
ARTN
|
artemin |
| chr17_+_29421900 | 0.30 |
ENST00000358273.4
ENST00000356175.3 |
NF1
|
neurofibromin 1 |
| chr14_+_57857262 | 0.30 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr3_-_176915036 | 0.30 |
ENST00000427349.1
ENST00000352800.5 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr18_-_29522989 | 0.29 |
ENST00000582539.1
ENST00000283351.4 ENST00000582513.1 |
TRAPPC8
|
trafficking protein particle complex 8 |
| chr1_-_46598371 | 0.28 |
ENST00000372006.1
ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr5_-_42811986 | 0.28 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr1_+_244816237 | 0.28 |
ENST00000302550.11
|
DESI2
|
desumoylating isopeptidase 2 |
| chr1_-_183604794 | 0.28 |
ENST00000367534.1
ENST00000359856.6 ENST00000294742.6 |
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa |
| chr20_-_62284766 | 0.28 |
ENST00000370053.1
|
STMN3
|
stathmin-like 3 |
| chr20_+_2083540 | 0.28 |
ENST00000400064.3
|
STK35
|
serine/threonine kinase 35 |
| chr11_+_117014983 | 0.27 |
ENST00000527958.1
ENST00000419197.2 ENST00000304808.6 ENST00000529887.2 |
PAFAH1B2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 (30kDa) |
| chr7_+_94139105 | 0.27 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr4_+_38665810 | 0.27 |
ENST00000261438.5
ENST00000514033.1 |
KLF3
|
Kruppel-like factor 3 (basic) |
| chr1_-_93426998 | 0.27 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr1_-_171711387 | 0.27 |
ENST00000236192.7
|
VAMP4
|
vesicle-associated membrane protein 4 |
| chr15_-_52970820 | 0.27 |
ENST00000261844.7
ENST00000399202.4 ENST00000562135.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr10_+_112404132 | 0.26 |
ENST00000369519.3
|
RBM20
|
RNA binding motif protein 20 |
| chr2_+_201170999 | 0.26 |
ENST00000439395.1
ENST00000444012.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr2_-_165698521 | 0.26 |
ENST00000409184.3
ENST00000392717.2 ENST00000456693.1 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr2_-_183903133 | 0.25 |
ENST00000361354.4
|
NCKAP1
|
NCK-associated protein 1 |
| chr1_+_95699704 | 0.25 |
ENST00000370202.4
|
RWDD3
|
RWD domain containing 3 |
| chrX_-_129244336 | 0.25 |
ENST00000434609.1
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr2_+_170683979 | 0.25 |
ENST00000418381.1
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr6_-_111804905 | 0.24 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr6_-_111804393 | 0.24 |
ENST00000368802.3
ENST00000368805.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr17_-_4269768 | 0.24 |
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr2_+_54198210 | 0.24 |
ENST00000607452.1
ENST00000422521.2 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr19_-_52097613 | 0.24 |
ENST00000301439.3
|
AC018755.1
|
HCG2008157; Uncharacterized protein; cDNA FLJ30403 fis, clone BRACE2008480 |
| chrX_-_119695279 | 0.24 |
ENST00000336592.6
|
CUL4B
|
cullin 4B |
| chr11_-_118023594 | 0.23 |
ENST00000529878.1
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr5_+_112312399 | 0.23 |
ENST00000515408.1
ENST00000513585.1 |
DCP2
|
decapping mRNA 2 |
| chr1_-_23810664 | 0.23 |
ENST00000336689.3
ENST00000437606.2 |
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr18_-_32924372 | 0.22 |
ENST00000261332.6
ENST00000399061.3 |
ZNF24
|
zinc finger protein 24 |
| chr2_-_60780607 | 0.22 |
ENST00000537768.1
ENST00000335712.6 ENST00000356842.4 |
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr2_+_201171577 | 0.22 |
ENST00000409397.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr6_+_56954867 | 0.22 |
ENST00000370708.4
ENST00000370702.1 |
ZNF451
|
zinc finger protein 451 |
| chr1_+_206138884 | 0.22 |
ENST00000341209.5
ENST00000607379.1 |
FAM72A
|
family with sequence similarity 72, member A |
| chr5_+_67511524 | 0.22 |
ENST00000521381.1
ENST00000521657.1 |
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr15_-_72490114 | 0.22 |
ENST00000309731.7
|
GRAMD2
|
GRAM domain containing 2 |
| chr1_+_235491714 | 0.21 |
ENST00000471812.1
ENST00000358966.2 ENST00000282841.5 ENST00000391855.2 |
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr20_+_49348081 | 0.21 |
ENST00000371610.2
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr5_-_137368726 | 0.21 |
ENST00000420893.2
ENST00000425075.2 |
FAM13B
|
family with sequence similarity 13, member B |
| chr20_+_49348109 | 0.21 |
ENST00000396039.1
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr2_-_38830160 | 0.21 |
ENST00000409636.1
ENST00000608859.1 ENST00000358367.4 |
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr17_+_42634844 | 0.21 |
ENST00000315323.3
|
FZD2
|
frizzled family receptor 2 |
| chr17_-_36413133 | 0.21 |
ENST00000523089.1
ENST00000312412.4 ENST00000520237.1 |
RP11-1407O15.2
|
TBC1 domain family member 3 |
| chr2_-_60780702 | 0.21 |
ENST00000359629.5
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr5_+_112312454 | 0.20 |
ENST00000543319.1
|
DCP2
|
decapping mRNA 2 |
| chr11_-_12030681 | 0.20 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr2_-_44223138 | 0.20 |
ENST00000260665.7
|
LRPPRC
|
leucine-rich pentatricopeptide repeat containing |
| chr13_-_96705624 | 0.20 |
ENST00000376747.3
ENST00000376712.4 ENST00000397618.3 ENST00000376714.3 |
UGGT2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr7_+_99156314 | 0.20 |
ENST00000425063.1
ENST00000493277.1 |
ZNF655
|
zinc finger protein 655 |
| chr4_-_120548779 | 0.19 |
ENST00000264805.5
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr1_+_180601139 | 0.19 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr10_-_69834973 | 0.19 |
ENST00000395187.2
|
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr18_-_51750948 | 0.19 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr1_-_159915386 | 0.19 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr12_+_57482665 | 0.19 |
ENST00000300131.3
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr6_+_126070726 | 0.19 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr11_-_117186946 | 0.18 |
ENST00000313005.6
ENST00000528053.1 |
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr6_-_110500826 | 0.18 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr3_-_129612394 | 0.18 |
ENST00000505616.1
ENST00000426664.2 |
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr2_+_14772810 | 0.18 |
ENST00000295092.2
ENST00000331243.4 |
FAM84A
|
family with sequence similarity 84, member A |
| chrX_-_131352152 | 0.18 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr22_-_24181174 | 0.18 |
ENST00000318109.7
ENST00000406855.3 ENST00000404056.1 ENST00000476077.1 |
DERL3
|
derlin 3 |
| chr10_-_112064665 | 0.18 |
ENST00000369603.5
|
SMNDC1
|
survival motor neuron domain containing 1 |
| chr3_-_100120223 | 0.17 |
ENST00000284320.5
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr16_+_2014941 | 0.17 |
ENST00000531523.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr8_-_67525473 | 0.17 |
ENST00000522677.3
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr3_-_11623804 | 0.17 |
ENST00000451674.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr8_-_27469383 | 0.17 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr5_+_61708582 | 0.17 |
ENST00000325324.6
|
IPO11
|
importin 11 |
| chr5_-_77590480 | 0.17 |
ENST00000519295.1
ENST00000255194.6 |
AP3B1
|
adaptor-related protein complex 3, beta 1 subunit |
| chr3_-_88108192 | 0.17 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr3_-_64431146 | 0.17 |
ENST00000569824.1
ENST00000485770.1 |
PRICKLE2
RP11-14D22.2
|
prickle homolog 2 (Drosophila) RP11-14D22.2 |
| chr13_+_25670268 | 0.16 |
ENST00000281589.3
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3 |
| chr12_+_109535373 | 0.16 |
ENST00000242576.2
|
UNG
|
uracil-DNA glycosylase |
| chrX_-_119763835 | 0.16 |
ENST00000371313.2
ENST00000304661.5 |
C1GALT1C1
|
C1GALT1-specific chaperone 1 |
| chr5_+_139028510 | 0.16 |
ENST00000502336.1
ENST00000520967.1 ENST00000511048.1 |
CXXC5
|
CXXC finger protein 5 |
| chr22_-_50218452 | 0.16 |
ENST00000216267.8
|
BRD1
|
bromodomain containing 1 |
| chr5_-_107717058 | 0.16 |
ENST00000359660.5
|
FBXL17
|
F-box and leucine-rich repeat protein 17 |
| chrX_+_123097014 | 0.16 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr2_+_32390925 | 0.16 |
ENST00000440718.1
ENST00000379343.2 ENST00000282587.5 ENST00000435660.1 ENST00000538303.1 ENST00000357055.3 ENST00000406369.1 |
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr2_+_26256938 | 0.16 |
ENST00000264710.4
|
RAB10
|
RAB10, member RAS oncogene family |
| chr22_-_50217981 | 0.16 |
ENST00000457780.2
|
BRD1
|
bromodomain containing 1 |
| chr1_-_236030216 | 0.16 |
ENST00000389794.3
ENST00000389793.2 |
LYST
|
lysosomal trafficking regulator |
| chr10_-_21463116 | 0.16 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr10_-_13570533 | 0.16 |
ENST00000396900.2
ENST00000396898.2 |
BEND7
|
BEN domain containing 7 |
| chr8_+_38614778 | 0.16 |
ENST00000521050.1
ENST00000522904.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr10_-_69835099 | 0.16 |
ENST00000373700.4
|
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr2_-_32390801 | 0.16 |
ENST00000608489.1
|
RP11-563N4.1
|
RP11-563N4.1 |
| chr3_+_170075436 | 0.15 |
ENST00000476188.1
ENST00000259119.4 ENST00000426052.2 |
SKIL
|
SKI-like oncogene |
| chr2_+_54342574 | 0.15 |
ENST00000303536.4
ENST00000394666.3 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr14_-_61190754 | 0.15 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr16_-_18937726 | 0.15 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr18_+_29672573 | 0.15 |
ENST00000578107.1
ENST00000257190.5 ENST00000580499.1 |
RNF138
|
ring finger protein 138, E3 ubiquitin protein ligase |
| chr19_-_49576198 | 0.15 |
ENST00000221444.1
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr1_+_226250379 | 0.15 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chr8_-_42396721 | 0.15 |
ENST00000518384.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr4_+_88928777 | 0.15 |
ENST00000237596.2
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant) |
| chr3_+_73045936 | 0.15 |
ENST00000356692.5
ENST00000488810.1 ENST00000394284.3 ENST00000295862.9 ENST00000495566.1 |
PPP4R2
|
protein phosphatase 4, regulatory subunit 2 |
| chr8_-_125486755 | 0.15 |
ENST00000499418.2
ENST00000530778.1 |
RNF139-AS1
|
RNF139 antisense RNA 1 (head to head) |
| chr19_-_10628117 | 0.14 |
ENST00000333430.4
|
S1PR5
|
sphingosine-1-phosphate receptor 5 |
| chrX_+_9754461 | 0.14 |
ENST00000380913.3
|
SHROOM2
|
shroom family member 2 |
| chr2_+_170683942 | 0.14 |
ENST00000272793.5
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr6_-_53530474 | 0.14 |
ENST00000370905.3
|
KLHL31
|
kelch-like family member 31 |
| chrX_+_133507327 | 0.14 |
ENST00000332070.3
ENST00000394292.1 ENST00000370799.1 ENST00000416404.2 |
PHF6
|
PHD finger protein 6 |
| chr12_-_95467267 | 0.14 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr12_+_56661033 | 0.14 |
ENST00000433805.2
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr2_-_38829768 | 0.14 |
ENST00000378915.3
|
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr16_-_28937027 | 0.14 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr11_-_12030746 | 0.14 |
ENST00000533813.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr6_+_64281906 | 0.14 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr13_+_98795664 | 0.14 |
ENST00000376581.5
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr5_-_142784003 | 0.14 |
ENST00000416954.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr1_-_197169672 | 0.14 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr8_+_38614754 | 0.13 |
ENST00000521642.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr12_+_41086215 | 0.13 |
ENST00000547702.1
ENST00000551424.1 |
CNTN1
|
contactin 1 |
| chr14_+_96968707 | 0.13 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr12_+_41086297 | 0.13 |
ENST00000551295.2
|
CNTN1
|
contactin 1 |
| chr13_-_52026730 | 0.13 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr5_+_112312416 | 0.13 |
ENST00000389063.2
|
DCP2
|
decapping mRNA 2 |
| chr11_+_57509020 | 0.13 |
ENST00000388857.4
ENST00000528798.1 |
C11orf31
|
chromosome 11 open reading frame 31 |
| chr8_+_38644715 | 0.13 |
ENST00000317827.4
ENST00000379931.3 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr12_+_27677085 | 0.13 |
ENST00000545334.1
ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr1_-_46598284 | 0.13 |
ENST00000423209.1
ENST00000262741.5 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr8_-_102217796 | 0.13 |
ENST00000519744.1
ENST00000311212.4 ENST00000521272.1 ENST00000519882.1 |
ZNF706
|
zinc finger protein 706 |
| chr3_+_111578027 | 0.12 |
ENST00000431670.2
ENST00000412622.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr9_-_134615326 | 0.12 |
ENST00000438647.1
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr15_-_51914810 | 0.12 |
ENST00000543779.2
ENST00000449909.3 |
DMXL2
|
Dmx-like 2 |
| chr4_-_151936416 | 0.12 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr19_+_48112371 | 0.12 |
ENST00000594866.1
|
GLTSCR1
|
glioma tumor suppressor candidate region gene 1 |
| chr3_+_111578640 | 0.12 |
ENST00000393925.3
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr10_+_70715884 | 0.12 |
ENST00000354185.4
|
DDX21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr19_-_15090488 | 0.12 |
ENST00000594383.1
ENST00000598504.1 ENST00000597262.1 |
SLC1A6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr5_-_142783694 | 0.12 |
ENST00000394466.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr13_-_52027134 | 0.12 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr3_-_176914963 | 0.12 |
ENST00000450267.1
ENST00000431674.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr10_+_5726764 | 0.12 |
ENST00000328090.5
ENST00000496681.1 |
FAM208B
|
family with sequence similarity 208, member B |
| chr11_+_32914579 | 0.12 |
ENST00000399302.2
|
QSER1
|
glutamine and serine rich 1 |
| chr2_-_60780536 | 0.12 |
ENST00000538214.1
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr9_-_94877658 | 0.12 |
ENST00000262554.2
ENST00000337841.4 |
SPTLC1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr20_-_5093713 | 0.12 |
ENST00000342308.5
ENST00000202834.7 |
TMEM230
|
transmembrane protein 230 |
| chr12_+_49212261 | 0.12 |
ENST00000547818.1
ENST00000547392.1 |
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr6_-_79787902 | 0.12 |
ENST00000275034.4
|
PHIP
|
pleckstrin homology domain interacting protein |
| chr5_+_159343688 | 0.11 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr9_+_79792269 | 0.11 |
ENST00000376634.4
ENST00000376636.3 ENST00000360280.3 |
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chr3_-_169381183 | 0.11 |
ENST00000494292.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr11_+_130184888 | 0.11 |
ENST00000602376.1
ENST00000532116.3 ENST00000602310.1 |
RP11-121M22.1
|
RP11-121M22.1 |
| chr11_-_12030629 | 0.11 |
ENST00000396505.2
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr1_+_235492300 | 0.11 |
ENST00000476121.1
ENST00000497327.1 |
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr2_-_201828356 | 0.11 |
ENST00000234296.2
|
ORC2
|
origin recognition complex, subunit 2 |
| chr8_-_27469196 | 0.11 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr8_+_38644778 | 0.11 |
ENST00000276520.8
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr2_-_44223089 | 0.11 |
ENST00000447246.1
ENST00000409946.1 ENST00000409659.1 |
LRPPRC
|
leucine-rich pentatricopeptide repeat containing |
| chr2_-_74405929 | 0.11 |
ENST00000396049.4
|
MOB1A
|
MOB kinase activator 1A |
| chr19_+_48673949 | 0.11 |
ENST00000328759.7
|
C19orf68
|
chromosome 19 open reading frame 68 |
| chr1_-_54303934 | 0.11 |
ENST00000537333.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr12_+_65672702 | 0.11 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr21_+_17102311 | 0.11 |
ENST00000285679.6
ENST00000351097.5 ENST00000285681.2 ENST00000400183.2 |
USP25
|
ubiquitin specific peptidase 25 |
| chr8_-_101734170 | 0.10 |
ENST00000522387.1
ENST00000518196.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_-_41131106 | 0.10 |
ENST00000372683.1
|
RIMS3
|
regulating synaptic membrane exocytosis 3 |
| chr3_-_33260707 | 0.10 |
ENST00000309558.3
|
SUSD5
|
sushi domain containing 5 |
| chr6_+_45390222 | 0.10 |
ENST00000359524.5
|
RUNX2
|
runt-related transcription factor 2 |
| chr1_-_87379785 | 0.10 |
ENST00000401030.3
ENST00000370554.1 |
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr12_+_110718428 | 0.10 |
ENST00000552636.1
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr7_-_48068643 | 0.09 |
ENST00000453192.2
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr17_+_46185111 | 0.09 |
ENST00000582104.1
ENST00000584335.1 |
SNX11
|
sorting nexin 11 |
| chr5_+_111496554 | 0.09 |
ENST00000442823.2
|
EPB41L4A-AS1
|
EPB41L4A antisense RNA 1 |
| chrX_+_133507283 | 0.09 |
ENST00000370803.3
|
PHF6
|
PHD finger protein 6 |
| chr1_-_109584716 | 0.09 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chr12_+_49212514 | 0.09 |
ENST00000301050.2
ENST00000548279.1 ENST00000547230.1 |
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr7_+_89841024 | 0.09 |
ENST00000394626.1
|
STEAP2
|
STEAP family member 2, metalloreductase |
| chr6_-_48036363 | 0.09 |
ENST00000543600.1
ENST00000398738.2 ENST00000339488.4 |
PTCHD4
|
patched domain containing 4 |
| chr11_+_117015024 | 0.09 |
ENST00000530272.1
|
PAFAH1B2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 (30kDa) |
| chr12_+_110719032 | 0.09 |
ENST00000395494.2
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr3_+_128720424 | 0.08 |
ENST00000480450.1
ENST00000436022.2 |
EFCC1
|
EF-hand and coiled-coil domain containing 1 |
| chr7_+_2559399 | 0.08 |
ENST00000222725.5
ENST00000359574.3 |
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_-_23670817 | 0.08 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr10_+_38299546 | 0.08 |
ENST00000374618.3
ENST00000432900.2 ENST00000458705.2 ENST00000469037.2 |
ZNF33A
|
zinc finger protein 33A |
| chr16_-_18937480 | 0.08 |
ENST00000532700.2
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR0B1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.2 | 0.6 | GO:1904799 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 | 0.4 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.5 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.3 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 0.3 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.1 | 0.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.3 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.2 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 | 0.2 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.1 | 0.2 | GO:0070632 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 0.2 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.1 | 0.2 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.1 | 0.5 | GO:1902613 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.1 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.2 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 | 0.4 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.3 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.6 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.0 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.5 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.2 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0060623 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.8 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.4 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.5 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.1 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.2 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.6 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0089712 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.0 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.3 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.6 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.0 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.2 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.1 | 0.5 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.5 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.2 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 1.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.3 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.2 | GO:0031775 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.1 | 0.4 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0048763 | HLH domain binding(GO:0043398) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.0 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.6 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |