Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
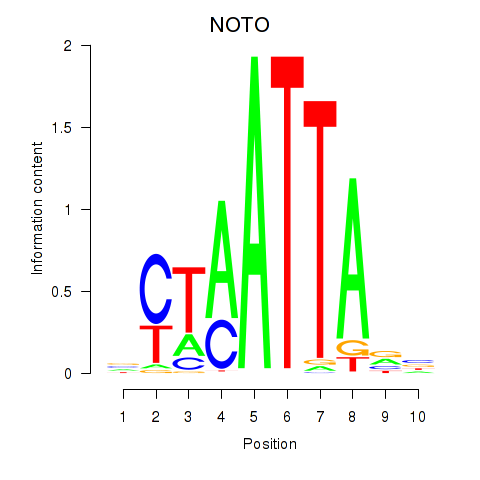
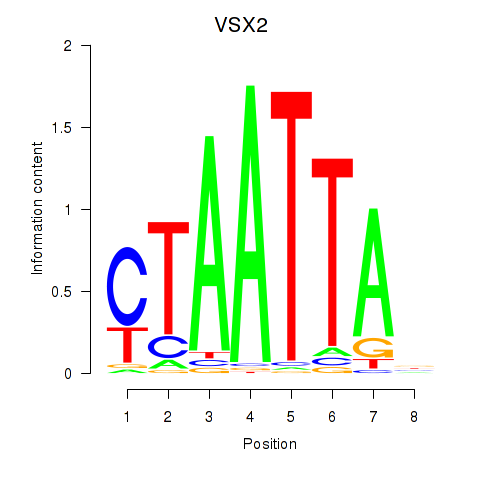
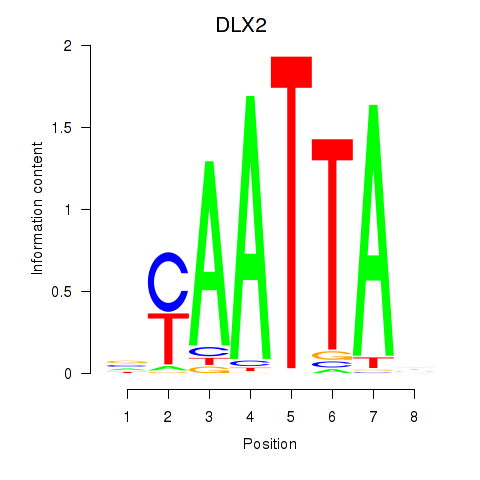
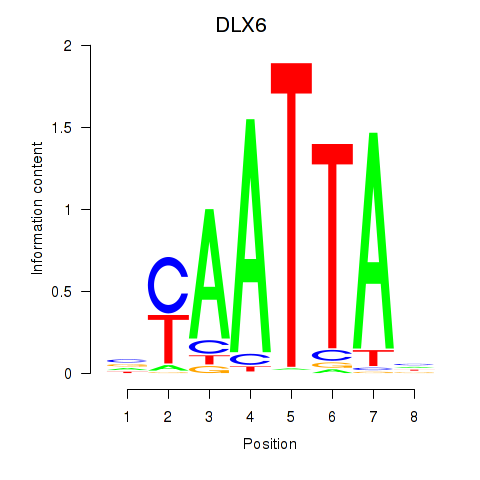
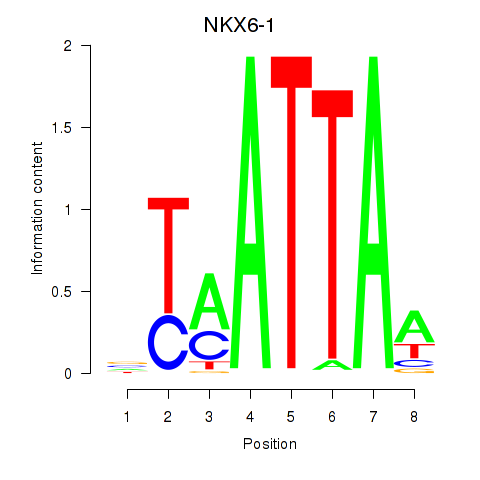
Results for NOTO_VSX2_DLX2_DLX6_NKX6-1
Z-value: 0.49
Transcription factors associated with NOTO_VSX2_DLX2_DLX6_NKX6-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NOTO
|
ENSG00000214513.3 | notochord homeobox |
|
VSX2
|
ENSG00000119614.2 | visual system homeobox 2 |
|
DLX2
|
ENSG00000115844.6 | distal-less homeobox 2 |
|
DLX6
|
ENSG00000006377.9 | distal-less homeobox 6 |
|
NKX6-1
|
ENSG00000163623.5 | NK6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DLX6 | hg19_v2_chr7_+_96634850_96634874 | 0.21 | 6.9e-01 | Click! |
| NOTO | hg19_v2_chr2_+_73429386_73429386 | 0.17 | 7.4e-01 | Click! |
| DLX2 | hg19_v2_chr2_-_172967621_172967637 | 0.01 | 9.9e-01 | Click! |
Activity profile of NOTO_VSX2_DLX2_DLX6_NKX6-1 motif
Sorted Z-values of NOTO_VSX2_DLX2_DLX6_NKX6-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_25281747 | 0.72 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr17_+_47448102 | 0.34 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr6_+_130339710 | 0.34 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr21_+_17909594 | 0.33 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_-_38928414 | 0.32 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr5_+_53751445 | 0.31 |
ENST00000302005.1
|
HSPB3
|
heat shock 27kDa protein 3 |
| chr8_-_72268721 | 0.28 |
ENST00000419131.1
ENST00000388743.2 |
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr5_+_36606700 | 0.26 |
ENST00000416645.2
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr8_+_105235572 | 0.25 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr4_+_71108300 | 0.22 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr14_-_92413727 | 0.22 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr3_-_141747950 | 0.22 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr6_-_39693111 | 0.22 |
ENST00000373215.3
ENST00000538893.1 ENST00000287152.7 ENST00000373216.3 |
KIF6
|
kinesin family member 6 |
| chr14_-_57197224 | 0.21 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr5_-_56778635 | 0.21 |
ENST00000423391.1
|
ACTBL2
|
actin, beta-like 2 |
| chr17_-_39123144 | 0.21 |
ENST00000355612.2
|
KRT39
|
keratin 39 |
| chr4_+_105828537 | 0.20 |
ENST00000515649.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr4_+_105828492 | 0.19 |
ENST00000506148.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr13_+_36050881 | 0.18 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr2_+_66918558 | 0.18 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chrX_-_133792480 | 0.18 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr20_-_50418972 | 0.18 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr21_+_17443521 | 0.17 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_-_184243561 | 0.17 |
ENST00000514470.1
ENST00000541814.1 |
CLDN24
|
claudin 24 |
| chr21_-_31538971 | 0.16 |
ENST00000286808.3
|
CLDN17
|
claudin 17 |
| chr17_+_59489112 | 0.16 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr8_-_72268889 | 0.16 |
ENST00000388742.4
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr7_-_111424462 | 0.15 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr13_-_36050819 | 0.15 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr20_-_50418947 | 0.15 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr4_+_26324474 | 0.15 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr13_-_41593425 | 0.14 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr15_+_93443419 | 0.14 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr5_-_88119580 | 0.12 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr11_+_35222629 | 0.12 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_+_115498761 | 0.12 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr21_+_17443434 | 0.12 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_-_140544386 | 0.12 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr5_-_126409159 | 0.11 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr11_-_129062093 | 0.11 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr12_-_28122980 | 0.11 |
ENST00000395868.3
ENST00000534890.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr5_-_24645078 | 0.11 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr9_-_75488984 | 0.11 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chrM_+_7586 | 0.11 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr4_-_138453606 | 0.11 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr14_+_61654271 | 0.11 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr1_+_68150744 | 0.11 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr6_+_155538093 | 0.11 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr14_+_59100774 | 0.10 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr17_-_38911580 | 0.10 |
ENST00000312150.4
|
KRT25
|
keratin 25 |
| chr6_-_31107127 | 0.10 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr10_+_123951957 | 0.10 |
ENST00000514539.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr6_+_139135648 | 0.10 |
ENST00000541398.1
|
ECT2L
|
epithelial cell transforming sequence 2 oncogene-like |
| chr7_+_66800928 | 0.10 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr12_-_28123206 | 0.10 |
ENST00000542963.1
ENST00000535992.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr3_-_131756559 | 0.10 |
ENST00000505957.1
|
CPNE4
|
copine IV |
| chr1_+_160160283 | 0.09 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr3_+_173116225 | 0.09 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr8_-_72268968 | 0.09 |
ENST00000388740.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr17_-_39623681 | 0.09 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr1_+_160160346 | 0.09 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr5_-_96478466 | 0.09 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr11_-_16419067 | 0.09 |
ENST00000533411.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr17_-_2117600 | 0.09 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr11_-_8290263 | 0.09 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr12_+_54378849 | 0.09 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr2_-_214017151 | 0.09 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr1_-_150738261 | 0.08 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr17_+_18086392 | 0.08 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr5_+_140625147 | 0.08 |
ENST00000231173.3
|
PCDHB15
|
protocadherin beta 15 |
| chr4_+_71384257 | 0.08 |
ENST00000339336.4
|
AMTN
|
amelotin |
| chr2_+_47454054 | 0.08 |
ENST00000426892.1
|
AC106869.2
|
AC106869.2 |
| chr13_-_36788718 | 0.08 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr8_-_10512569 | 0.08 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr4_-_186696425 | 0.08 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_-_88120083 | 0.08 |
ENST00000509373.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr10_+_118083919 | 0.08 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chr12_+_54378923 | 0.08 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr1_+_225600404 | 0.08 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr19_-_12889226 | 0.08 |
ENST00000589400.1
ENST00000590839.1 ENST00000592079.1 |
HOOK2
|
hook microtubule-tethering protein 2 |
| chr1_-_92952433 | 0.08 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr7_+_99717230 | 0.08 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr20_+_52105495 | 0.08 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr2_+_190541153 | 0.08 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chrM_+_10758 | 0.07 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr3_-_71777824 | 0.07 |
ENST00000469524.1
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr3_+_111718173 | 0.07 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr1_+_183774285 | 0.07 |
ENST00000539189.1
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr4_-_89442940 | 0.07 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr21_+_17792672 | 0.07 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr7_-_14028488 | 0.07 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr1_-_68698222 | 0.07 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr3_-_126327398 | 0.07 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr5_+_147582387 | 0.07 |
ENST00000325630.2
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr20_-_60294804 | 0.07 |
ENST00000317652.1
|
RP11-429E11.3
|
Uncharacterized protein |
| chr17_+_43238438 | 0.07 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chrX_+_68835911 | 0.07 |
ENST00000525810.1
ENST00000527388.1 ENST00000374553.2 ENST00000374552.4 ENST00000338901.3 ENST00000524573.1 |
EDA
|
ectodysplasin A |
| chr9_+_82188077 | 0.07 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr18_-_53089723 | 0.07 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr1_-_68698197 | 0.07 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr20_-_50419055 | 0.07 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr5_+_60933634 | 0.07 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr14_-_36988882 | 0.07 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr15_+_67841330 | 0.07 |
ENST00000354498.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr6_-_33160231 | 0.06 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr5_-_124082279 | 0.06 |
ENST00000513986.1
|
ZNF608
|
zinc finger protein 608 |
| chr19_-_7698599 | 0.06 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr17_-_78450398 | 0.06 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr13_-_31040060 | 0.06 |
ENST00000326004.4
ENST00000341423.5 |
HMGB1
|
high mobility group box 1 |
| chr15_+_76352178 | 0.06 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr17_-_39203519 | 0.06 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chrX_-_46187069 | 0.06 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr2_-_182545603 | 0.06 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr12_-_6233828 | 0.06 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr1_+_61548374 | 0.06 |
ENST00000485903.2
ENST00000371185.2 ENST00000371184.2 |
NFIA
|
nuclear factor I/A |
| chr7_+_139528952 | 0.06 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr14_+_72052983 | 0.06 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr15_+_67390920 | 0.06 |
ENST00000559092.1
ENST00000560175.1 |
SMAD3
|
SMAD family member 3 |
| chr3_-_178984759 | 0.06 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr12_-_30887948 | 0.06 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr6_+_4087664 | 0.06 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chrX_-_64196351 | 0.06 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr1_-_45956868 | 0.06 |
ENST00000451835.2
|
TESK2
|
testis-specific kinase 2 |
| chr7_-_14026123 | 0.06 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr13_-_95131923 | 0.06 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chrM_-_14670 | 0.06 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr10_+_24755416 | 0.06 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr20_+_42187682 | 0.05 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chrX_-_73512358 | 0.05 |
ENST00000602776.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chrX_+_13671225 | 0.05 |
ENST00000545566.1
ENST00000544987.1 ENST00000314720.4 |
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr2_-_224467093 | 0.05 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr12_-_16760021 | 0.05 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr3_+_159557637 | 0.05 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr14_-_21994337 | 0.05 |
ENST00000537235.1
ENST00000450879.2 |
SALL2
|
spalt-like transcription factor 2 |
| chr4_+_169418255 | 0.05 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr17_+_72931876 | 0.05 |
ENST00000328801.4
|
OTOP3
|
otopetrin 3 |
| chr14_-_92413353 | 0.05 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr9_+_82267508 | 0.05 |
ENST00000490347.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr15_-_37393406 | 0.05 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr2_-_203735586 | 0.05 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr6_-_30899924 | 0.05 |
ENST00000359086.3
|
SFTA2
|
surfactant associated 2 |
| chr6_-_26235206 | 0.05 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr1_-_9953295 | 0.05 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr5_-_20575959 | 0.05 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr1_-_152131703 | 0.05 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr16_+_24621546 | 0.05 |
ENST00000566108.1
|
CTD-2540M10.1
|
CTD-2540M10.1 |
| chr6_+_5261225 | 0.05 |
ENST00000324331.6
|
FARS2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr17_-_39140549 | 0.05 |
ENST00000377755.4
|
KRT40
|
keratin 40 |
| chr4_-_177116772 | 0.05 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr12_-_54653313 | 0.05 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr10_+_4828815 | 0.05 |
ENST00000533295.1
|
AKR1E2
|
aldo-keto reductase family 1, member E2 |
| chr7_-_111032971 | 0.05 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr7_-_130080977 | 0.05 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr2_-_227050079 | 0.05 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chrM_+_14741 | 0.05 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr2_-_40680578 | 0.05 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr10_-_96829246 | 0.05 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr8_-_101661887 | 0.05 |
ENST00000311812.2
|
SNX31
|
sorting nexin 31 |
| chr12_-_15114658 | 0.05 |
ENST00000542276.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr17_-_40337470 | 0.05 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr4_-_186696561 | 0.05 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr20_+_56136136 | 0.05 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr6_-_5261141 | 0.05 |
ENST00000330636.4
ENST00000500576.2 |
LYRM4
|
LYR motif containing 4 |
| chr12_+_25205628 | 0.05 |
ENST00000554942.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr16_+_69345243 | 0.05 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chrM_+_10464 | 0.05 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr5_+_31193847 | 0.05 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr17_-_61045902 | 0.05 |
ENST00000581596.1
|
RP11-180P8.3
|
RP11-180P8.3 |
| chrX_-_106243451 | 0.05 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr11_+_34663913 | 0.04 |
ENST00000532302.1
|
EHF
|
ets homologous factor |
| chr7_-_14029515 | 0.04 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr11_-_107729287 | 0.04 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr7_-_14026063 | 0.04 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr15_+_45544426 | 0.04 |
ENST00000347644.3
ENST00000560438.1 |
SLC28A2
|
solute carrier family 28 (concentrative nucleoside transporter), member 2 |
| chr11_-_124981475 | 0.04 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chrX_+_100224676 | 0.04 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr6_+_148593425 | 0.04 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr12_-_56321397 | 0.04 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr3_-_101039402 | 0.04 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr3_+_156799587 | 0.04 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr12_+_25205155 | 0.04 |
ENST00000550945.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr12_-_53994805 | 0.04 |
ENST00000328463.7
|
ATF7
|
activating transcription factor 7 |
| chr3_+_15045419 | 0.04 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr3_-_120400960 | 0.04 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr1_+_214161272 | 0.04 |
ENST00000498508.2
ENST00000366958.4 |
PROX1
|
prospero homeobox 1 |
| chrM_+_3299 | 0.04 |
ENST00000361390.2
|
MT-ND1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr2_-_70780770 | 0.04 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr6_-_123958051 | 0.04 |
ENST00000546248.1
|
TRDN
|
triadin |
| chr9_+_44867571 | 0.04 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr1_+_168250194 | 0.04 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr17_+_48243352 | 0.04 |
ENST00000344627.6
ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA
|
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr6_-_111136299 | 0.04 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr3_-_114173530 | 0.04 |
ENST00000470311.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr17_+_46970127 | 0.04 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr17_+_46970134 | 0.04 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr7_-_84121858 | 0.04 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr20_+_58630972 | 0.04 |
ENST00000313426.1
|
C20orf197
|
chromosome 20 open reading frame 197 |
| chr5_+_147582348 | 0.04 |
ENST00000514389.1
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr6_+_151646800 | 0.04 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr8_-_72274095 | 0.04 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of NOTO_VSX2_DLX2_DLX6_NKX6-1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.6 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.0 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.3 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.1 | GO:2000979 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) negative regulation of bile acid biosynthetic process(GO:0070858) acinar cell differentiation(GO:0090425) negative regulation of bile acid metabolic process(GO:1904252) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.1 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.0 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.0 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.0 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.0 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.0 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |