Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
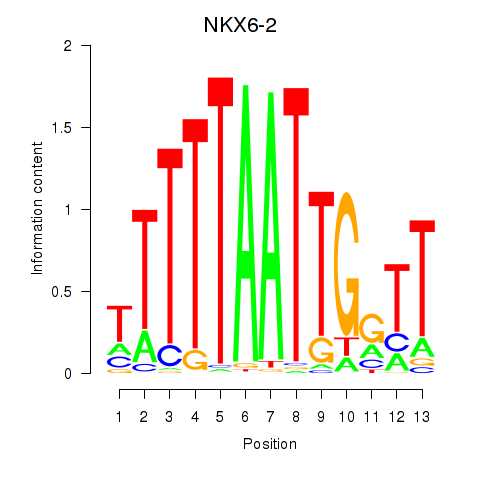
Results for NKX6-2
Z-value: 0.95
Transcription factors associated with NKX6-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX6-2
|
ENSG00000148826.6 | NK6 homeobox 2 |
Activity profile of NKX6-2 motif
Sorted Z-values of NKX6-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_23891693 | 1.05 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr8_+_92114873 | 0.88 |
ENST00000343709.3
ENST00000448384.2 |
LRRC69
|
leucine rich repeat containing 69 |
| chr6_-_113953705 | 0.69 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr7_-_77325545 | 0.61 |
ENST00000447009.1
ENST00000416650.1 ENST00000440088.1 ENST00000430801.1 ENST00000398043.2 |
RSBN1L-AS1
|
RSBN1L antisense RNA 1 |
| chr6_+_41021027 | 0.58 |
ENST00000244669.2
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr7_-_84122033 | 0.58 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chrX_-_80457385 | 0.52 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr15_+_49715449 | 0.49 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr9_+_109685630 | 0.47 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr18_+_76829441 | 0.44 |
ENST00000458297.2
|
ATP9B
|
ATPase, class II, type 9B |
| chr13_-_45048386 | 0.41 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr16_+_70695570 | 0.40 |
ENST00000597002.1
|
FLJ00418
|
FLJ00418 |
| chr1_+_85527987 | 0.39 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr14_+_86401039 | 0.39 |
ENST00000557195.1
|
CTD-2341M24.1
|
CTD-2341M24.1 |
| chr2_+_211342400 | 0.39 |
ENST00000417946.1
ENST00000518043.1 ENST00000523702.1 |
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr1_-_221915418 | 0.38 |
ENST00000323825.3
ENST00000366899.3 |
DUSP10
|
dual specificity phosphatase 10 |
| chr12_+_4385230 | 0.38 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chr2_-_136678123 | 0.37 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr12_+_25205568 | 0.37 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr22_+_46476192 | 0.35 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chr14_-_51027838 | 0.35 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr18_+_76829385 | 0.34 |
ENST00000426216.2
ENST00000307671.7 ENST00000586672.1 ENST00000586722.1 |
ATP9B
|
ATPase, class II, type 9B |
| chr12_+_41136144 | 0.33 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr15_+_49715293 | 0.32 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr8_-_42358742 | 0.31 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr8_+_79428539 | 0.30 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr5_-_111091948 | 0.30 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr7_+_77469439 | 0.29 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr8_+_104831472 | 0.29 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr8_+_97597148 | 0.28 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr1_+_78383813 | 0.27 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr2_+_172309634 | 0.27 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr17_+_57233087 | 0.27 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr12_+_32655048 | 0.26 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_-_211342292 | 0.26 |
ENST00000448951.1
|
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial) |
| chr1_-_110933611 | 0.26 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr6_-_26199471 | 0.25 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr3_+_156799587 | 0.25 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr1_-_110933663 | 0.25 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr5_+_95066823 | 0.25 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr15_-_49103184 | 0.24 |
ENST00000399334.3
ENST00000325747.5 |
CEP152
|
centrosomal protein 152kDa |
| chr6_-_56716686 | 0.24 |
ENST00000520645.1
|
DST
|
dystonin |
| chr2_+_210444298 | 0.24 |
ENST00000445941.1
|
MAP2
|
microtubule-associated protein 2 |
| chr7_-_22233442 | 0.23 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr14_+_76776957 | 0.23 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr1_-_109935819 | 0.23 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr6_-_53481847 | 0.22 |
ENST00000503985.1
|
RP1-27K12.2
|
RP1-27K12.2 |
| chr8_+_42873548 | 0.22 |
ENST00000533338.1
ENST00000534420.1 |
HOOK3
RP11-598P20.5
|
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr14_+_52164820 | 0.22 |
ENST00000554167.1
|
FRMD6
|
FERM domain containing 6 |
| chrX_+_105937068 | 0.22 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chrX_+_55744228 | 0.21 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr6_+_114178512 | 0.21 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr11_-_62521614 | 0.21 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr9_+_34652164 | 0.21 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr16_+_89228757 | 0.21 |
ENST00000565008.1
|
LINC00304
|
long intergenic non-protein coding RNA 304 |
| chr1_+_32379174 | 0.20 |
ENST00000391369.1
|
AL136115.1
|
HCG2032337; PRO1848; Uncharacterized protein |
| chr2_+_179149636 | 0.20 |
ENST00000409631.1
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr6_-_80247140 | 0.20 |
ENST00000392959.1
ENST00000467898.3 |
LCA5
|
Leber congenital amaurosis 5 |
| chr7_-_92146729 | 0.20 |
ENST00000541751.1
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr10_-_115904361 | 0.20 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr8_+_26150628 | 0.19 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr15_-_49103235 | 0.19 |
ENST00000380950.2
|
CEP152
|
centrosomal protein 152kDa |
| chr5_+_7373230 | 0.19 |
ENST00000500616.2
|
CTD-2296D1.5
|
CTD-2296D1.5 |
| chr4_+_95174445 | 0.19 |
ENST00000509418.1
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr6_+_42749759 | 0.19 |
ENST00000314073.5
|
GLTSCR1L
|
GLTSCR1-like |
| chr9_-_70465758 | 0.19 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chrX_+_55744166 | 0.19 |
ENST00000374941.4
ENST00000414239.1 |
RRAGB
|
Ras-related GTP binding B |
| chr6_-_80247105 | 0.18 |
ENST00000369846.4
|
LCA5
|
Leber congenital amaurosis 5 |
| chr8_-_72268721 | 0.18 |
ENST00000419131.1
ENST00000388743.2 |
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr16_+_53483983 | 0.18 |
ENST00000544545.1
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr9_+_88556036 | 0.17 |
ENST00000361671.5
ENST00000416045.1 |
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr12_-_76462713 | 0.17 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr6_+_47666275 | 0.17 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr10_+_6625605 | 0.17 |
ENST00000414894.1
ENST00000449648.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr1_+_210501589 | 0.17 |
ENST00000413764.2
ENST00000541565.1 |
HHAT
|
hedgehog acyltransferase |
| chr13_+_27844464 | 0.17 |
ENST00000241463.4
|
RASL11A
|
RAS-like, family 11, member A |
| chrX_+_105936982 | 0.17 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr12_+_111284764 | 0.16 |
ENST00000545036.1
ENST00000308208.5 |
CCDC63
|
coiled-coil domain containing 63 |
| chr5_+_67586465 | 0.16 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr8_-_103876965 | 0.16 |
ENST00000337198.5
|
AZIN1
|
antizyme inhibitor 1 |
| chr17_-_40829026 | 0.16 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr17_-_57232525 | 0.16 |
ENST00000583380.1
ENST00000580541.1 ENST00000578105.1 ENST00000437036.2 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr2_-_208030886 | 0.16 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr2_+_200472779 | 0.16 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chr9_-_77643307 | 0.15 |
ENST00000376834.3
ENST00000376830.3 |
C9orf41
|
chromosome 9 open reading frame 41 |
| chrX_+_102840408 | 0.15 |
ENST00000468024.1
ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4
|
transcription elongation factor A (SII)-like 4 |
| chr2_-_191115229 | 0.15 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr18_+_61575200 | 0.15 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr17_-_40828969 | 0.15 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr10_-_47239738 | 0.14 |
ENST00000413193.2
|
AGAP10
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr12_+_40787194 | 0.14 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr14_+_63671105 | 0.14 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr12_-_74686314 | 0.14 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr9_-_110540419 | 0.14 |
ENST00000398726.3
|
AL162389.1
|
Uncharacterized protein |
| chr13_+_97928395 | 0.13 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr4_-_25865159 | 0.13 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr12_+_32655110 | 0.13 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr18_-_53019208 | 0.13 |
ENST00000562607.1
|
TCF4
|
transcription factor 4 |
| chr2_+_210443993 | 0.13 |
ENST00000392193.1
|
MAP2
|
microtubule-associated protein 2 |
| chr5_+_68513622 | 0.13 |
ENST00000512880.1
ENST00000602380.1 |
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr8_+_92114060 | 0.13 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr2_+_218989991 | 0.13 |
ENST00000453237.1
|
CXCR2
|
chemokine (C-X-C motif) receptor 2 |
| chr22_-_40929812 | 0.12 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr7_-_20256965 | 0.12 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr1_+_174670143 | 0.12 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr1_-_32384693 | 0.12 |
ENST00000602683.1
ENST00000470404.1 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr19_+_52772821 | 0.12 |
ENST00000439461.1
|
ZNF766
|
zinc finger protein 766 |
| chr18_-_53255379 | 0.11 |
ENST00000565908.2
|
TCF4
|
transcription factor 4 |
| chr6_+_25963020 | 0.11 |
ENST00000357085.3
|
TRIM38
|
tripartite motif containing 38 |
| chr16_+_28565230 | 0.11 |
ENST00000317058.3
|
CCDC101
|
coiled-coil domain containing 101 |
| chr6_+_116850174 | 0.11 |
ENST00000416171.2
ENST00000368597.2 ENST00000452373.1 ENST00000405399.1 |
FAM26D
|
family with sequence similarity 26, member D |
| chr4_+_169013666 | 0.11 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr17_-_57232596 | 0.10 |
ENST00000581068.1
ENST00000330137.7 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr17_-_56494882 | 0.10 |
ENST00000584437.1
|
RNF43
|
ring finger protein 43 |
| chr6_-_160209471 | 0.10 |
ENST00000539948.1
|
TCP1
|
t-complex 1 |
| chr21_-_36421535 | 0.10 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chr1_+_145477060 | 0.10 |
ENST00000369308.3
|
LIX1L
|
Lix1 homolog (mouse)-like |
| chr12_+_28410128 | 0.10 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr1_+_244624678 | 0.10 |
ENST00000366534.4
ENST00000366533.4 ENST00000428042.1 ENST00000366531.3 |
C1orf101
|
chromosome 1 open reading frame 101 |
| chr2_-_74570520 | 0.10 |
ENST00000394019.2
ENST00000346834.4 ENST00000359484.4 ENST00000423644.1 ENST00000377634.4 ENST00000436454.1 |
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr5_-_159546396 | 0.10 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr10_-_63995871 | 0.09 |
ENST00000315289.2
|
RTKN2
|
rhotekin 2 |
| chr19_+_52772832 | 0.09 |
ENST00000593703.1
ENST00000601711.1 ENST00000599581.1 |
ZNF766
|
zinc finger protein 766 |
| chr21_-_36421626 | 0.09 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chr1_-_200379180 | 0.09 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr14_-_98444438 | 0.09 |
ENST00000512901.2
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr19_+_12175504 | 0.09 |
ENST00000439326.3
|
ZNF844
|
zinc finger protein 844 |
| chr21_-_36421401 | 0.09 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr2_+_179318295 | 0.09 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr1_+_149858461 | 0.09 |
ENST00000331380.2
|
HIST2H2AC
|
histone cluster 2, H2ac |
| chr4_-_87374283 | 0.09 |
ENST00000361569.2
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr9_-_26892342 | 0.09 |
ENST00000535437.1
|
CAAP1
|
caspase activity and apoptosis inhibitor 1 |
| chr6_-_116833500 | 0.09 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chrX_-_10544942 | 0.08 |
ENST00000380779.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr16_+_21610879 | 0.08 |
ENST00000396014.4
|
METTL9
|
methyltransferase like 9 |
| chr7_+_77325738 | 0.08 |
ENST00000334955.8
|
RSBN1L
|
round spermatid basic protein 1-like |
| chr3_-_45837959 | 0.08 |
ENST00000353278.4
ENST00000456124.2 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr2_+_210444142 | 0.08 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr10_+_123951957 | 0.08 |
ENST00000514539.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr7_-_111032971 | 0.08 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr1_+_196621002 | 0.08 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr12_+_111284805 | 0.07 |
ENST00000552694.1
|
CCDC63
|
coiled-coil domain containing 63 |
| chr7_-_115799942 | 0.07 |
ENST00000484212.1
|
TFEC
|
transcription factor EC |
| chr2_-_65593784 | 0.07 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr16_+_14841923 | 0.07 |
ENST00000530328.1
ENST00000529166.1 |
NPIPA2
|
nuclear pore complex interacting protein family, member A2 |
| chr20_+_57467204 | 0.07 |
ENST00000603546.1
|
GNAS
|
GNAS complex locus |
| chr1_+_100810575 | 0.07 |
ENST00000542213.1
|
CDC14A
|
cell division cycle 14A |
| chr18_-_47018897 | 0.07 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr2_+_108994466 | 0.07 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr10_-_14574705 | 0.07 |
ENST00000489100.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr14_-_98444369 | 0.07 |
ENST00000554822.1
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr1_+_180601139 | 0.07 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr22_+_24198890 | 0.07 |
ENST00000345044.6
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr1_+_174933899 | 0.07 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr1_+_152178320 | 0.07 |
ENST00000429352.1
|
RP11-107M16.2
|
RP11-107M16.2 |
| chr18_-_47018769 | 0.07 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr18_-_52989525 | 0.07 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr2_+_202047843 | 0.06 |
ENST00000272879.5
ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr5_-_78365437 | 0.06 |
ENST00000380311.4
ENST00000540686.1 ENST00000255189.3 |
DMGDH
|
dimethylglycine dehydrogenase |
| chr8_+_104831440 | 0.06 |
ENST00000515551.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr18_+_32290218 | 0.06 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr5_+_68513557 | 0.06 |
ENST00000256441.4
|
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr3_-_45838011 | 0.06 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr15_+_80364901 | 0.06 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr2_-_207024134 | 0.06 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr2_-_217560248 | 0.06 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr9_-_23825956 | 0.06 |
ENST00000397312.2
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr4_-_108204846 | 0.06 |
ENST00000513208.1
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr11_-_11374904 | 0.06 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr2_+_118572226 | 0.06 |
ENST00000263239.2
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr17_+_72581057 | 0.06 |
ENST00000392620.1
|
C17orf77
|
chromosome 17 open reading frame 77 |
| chr10_-_71169031 | 0.05 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr14_+_73706308 | 0.05 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr1_-_149858227 | 0.05 |
ENST00000369155.2
|
HIST2H2BE
|
histone cluster 2, H2be |
| chr20_-_18477862 | 0.05 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr12_+_14561422 | 0.05 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr6_-_26199499 | 0.05 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr18_-_47018869 | 0.05 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr16_+_14802801 | 0.05 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr4_+_55095428 | 0.05 |
ENST00000508170.1
ENST00000512143.1 |
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr3_+_16216137 | 0.04 |
ENST00000339732.5
|
GALNT15
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chr10_+_6625733 | 0.04 |
ENST00000607982.1
ENST00000608526.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr11_+_57529234 | 0.04 |
ENST00000360682.6
ENST00000361796.4 ENST00000529526.1 ENST00000426142.2 ENST00000399050.4 ENST00000361391.6 ENST00000361332.4 ENST00000532463.1 ENST00000529986.1 ENST00000358694.6 ENST00000532787.1 ENST00000533667.1 ENST00000532649.1 ENST00000528621.1 ENST00000530748.1 ENST00000428599.2 ENST00000527467.1 ENST00000528232.1 ENST00000531014.1 ENST00000526772.1 ENST00000529873.1 ENST00000525902.1 ENST00000532844.1 ENST00000526357.1 ENST00000530094.1 ENST00000415361.2 ENST00000532245.1 ENST00000534579.1 ENST00000526938.1 |
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr2_-_207023918 | 0.04 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr9_-_23826298 | 0.04 |
ENST00000380117.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr14_-_99737565 | 0.04 |
ENST00000357195.3
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr2_-_207024233 | 0.04 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr11_-_31531121 | 0.04 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr10_-_94257512 | 0.04 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme |
| chr14_-_27066636 | 0.04 |
ENST00000267422.7
ENST00000344429.5 ENST00000574031.1 ENST00000465357.2 ENST00000547619.1 |
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr8_-_145754428 | 0.03 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr18_-_3220106 | 0.03 |
ENST00000356443.4
ENST00000400569.3 |
MYOM1
|
myomesin 1 |
| chr6_-_64029879 | 0.03 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr1_-_174992544 | 0.03 |
ENST00000476371.1
|
MRPS14
|
mitochondrial ribosomal protein S14 |
| chr16_+_19422035 | 0.03 |
ENST00000381414.4
ENST00000396229.2 |
TMC5
|
transmembrane channel-like 5 |
| chr4_+_26324474 | 0.03 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr7_-_140482926 | 0.03 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr3_-_141747459 | 0.03 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr22_-_29137771 | 0.03 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr18_+_72922710 | 0.03 |
ENST00000322038.5
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX6-2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of mitochondrial DNA metabolic process(GO:1901860) response to methionine(GO:1904640) |
| 0.2 | 0.8 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.6 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.4 | GO:0070408 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.1 | 0.4 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.4 | GO:0060264 | respiratory burst involved in inflammatory response(GO:0002536) regulation of respiratory burst involved in inflammatory response(GO:0060264) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.6 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.2 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.4 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.4 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0051808 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:1904204 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) operant conditioning(GO:0035106) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.4 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.5 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.0 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0072501 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 1.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.4 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 1.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.0 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0046935 | ErbB-3 class receptor binding(GO:0043125) 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |