Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
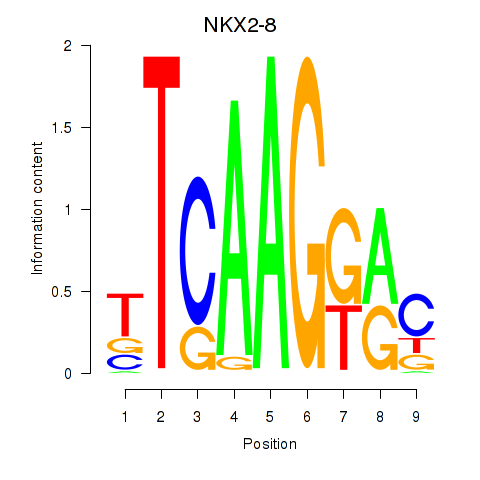
Results for NKX2-8
Z-value: 0.68
Transcription factors associated with NKX2-8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-8
|
ENSG00000136327.6 | NK2 homeobox 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-8 | hg19_v2_chr14_-_37051798_37051831 | -0.15 | 7.7e-01 | Click! |
Activity profile of NKX2-8 motif
Sorted Z-values of NKX2-8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_27998689 | 0.82 |
ENST00000339145.4
ENST00000362020.4 ENST00000361157.6 |
IFI6
|
interferon, alpha-inducible protein 6 |
| chr2_-_25451065 | 0.53 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr17_+_67590125 | 0.53 |
ENST00000591334.1
|
AC003051.1
|
AC003051.1 |
| chrX_+_115567767 | 0.42 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr5_-_176056974 | 0.39 |
ENST00000510387.1
ENST00000506696.1 |
SNCB
|
synuclein, beta |
| chr1_+_10057274 | 0.38 |
ENST00000294435.7
|
RBP7
|
retinol binding protein 7, cellular |
| chr3_+_177545563 | 0.32 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr22_+_38609538 | 0.31 |
ENST00000407965.1
|
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr6_+_43968306 | 0.31 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr14_-_24732403 | 0.28 |
ENST00000206765.6
|
TGM1
|
transglutaminase 1 |
| chr2_-_36779411 | 0.28 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr3_+_172361483 | 0.27 |
ENST00000598405.1
|
AC007919.2
|
HCG1787166; PRO1163; Uncharacterized protein |
| chr16_+_23194033 | 0.25 |
ENST00000300061.2
|
SCNN1G
|
sodium channel, non-voltage-gated 1, gamma subunit |
| chr6_+_41010293 | 0.25 |
ENST00000373161.1
ENST00000373158.2 ENST00000470917.1 |
TSPO2
|
translocator protein 2 |
| chr2_+_119699864 | 0.23 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr17_+_76210367 | 0.22 |
ENST00000592734.1
ENST00000587746.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr2_-_159313214 | 0.20 |
ENST00000409889.1
ENST00000283233.5 ENST00000536771.1 |
CCDC148
|
coiled-coil domain containing 148 |
| chr3_+_126423045 | 0.20 |
ENST00000290913.3
ENST00000508789.1 |
CHCHD6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr14_-_24732368 | 0.20 |
ENST00000544573.1
|
TGM1
|
transglutaminase 1 |
| chr7_-_127671674 | 0.19 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr12_+_49658855 | 0.19 |
ENST00000549183.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr2_+_119699742 | 0.17 |
ENST00000327097.4
|
MARCO
|
macrophage receptor with collagenous structure |
| chr8_+_107593198 | 0.17 |
ENST00000517686.1
|
OXR1
|
oxidation resistance 1 |
| chr14_+_58894404 | 0.17 |
ENST00000554463.1
ENST00000555833.1 |
KIAA0586
|
KIAA0586 |
| chr11_-_128457446 | 0.16 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr3_-_16524357 | 0.16 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr1_-_95007193 | 0.16 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr1_+_228337553 | 0.15 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr2_+_101591314 | 0.15 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr2_-_68547061 | 0.15 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr4_+_76481258 | 0.15 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr21_+_33671160 | 0.15 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr3_+_239652 | 0.15 |
ENST00000435603.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr18_-_72920372 | 0.14 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr12_+_13349711 | 0.14 |
ENST00000538364.1
ENST00000396301.3 |
EMP1
|
epithelial membrane protein 1 |
| chr7_-_766879 | 0.14 |
ENST00000537384.1
ENST00000417852.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr12_+_66218903 | 0.14 |
ENST00000393577.3
|
HMGA2
|
high mobility group AT-hook 2 |
| chr8_+_105602961 | 0.13 |
ENST00000521923.1
|
RP11-127H5.1
|
Uncharacterized protein |
| chr3_+_67048721 | 0.13 |
ENST00000295568.4
ENST00000484414.1 ENST00000460576.1 ENST00000417314.2 |
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr18_+_44497455 | 0.13 |
ENST00000592005.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr17_+_61086917 | 0.13 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chrX_+_48681768 | 0.12 |
ENST00000430858.1
|
HDAC6
|
histone deacetylase 6 |
| chr22_+_39101728 | 0.12 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr8_-_29120604 | 0.12 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr15_-_60683326 | 0.12 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr22_-_32341336 | 0.11 |
ENST00000248984.3
|
C22orf24
|
chromosome 22 open reading frame 24 |
| chr15_-_35047166 | 0.10 |
ENST00000290374.4
|
GJD2
|
gap junction protein, delta 2, 36kDa |
| chr12_+_7941989 | 0.10 |
ENST00000229307.4
|
NANOG
|
Nanog homeobox |
| chr4_-_100485143 | 0.10 |
ENST00000394877.3
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr8_-_29120580 | 0.10 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr16_+_58426296 | 0.10 |
ENST00000426538.2
ENST00000328514.7 ENST00000318129.5 |
GINS3
|
GINS complex subunit 3 (Psf3 homolog) |
| chrM_+_8366 | 0.10 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chrX_-_69479654 | 0.10 |
ENST00000374519.2
|
P2RY4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr19_-_39881669 | 0.10 |
ENST00000221266.7
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr10_-_135171178 | 0.10 |
ENST00000368551.1
|
FUOM
|
fucose mutarotase |
| chr18_+_3450161 | 0.09 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr1_+_180165672 | 0.09 |
ENST00000443059.1
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr5_-_95158644 | 0.09 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr21_+_33671264 | 0.09 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr3_-_52739670 | 0.09 |
ENST00000497953.1
|
GLT8D1
|
glycosyltransferase 8 domain containing 1 |
| chr3_-_114790179 | 0.09 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr7_-_95025661 | 0.09 |
ENST00000542556.1
ENST00000265627.5 ENST00000427422.1 ENST00000451904.1 |
PON1
PON3
|
paraoxonase 1 paraoxonase 3 |
| chr11_+_59705928 | 0.09 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr20_-_48747662 | 0.09 |
ENST00000371656.2
|
TMEM189
|
transmembrane protein 189 |
| chr18_+_13382553 | 0.09 |
ENST00000586222.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr8_+_23430157 | 0.09 |
ENST00000399967.3
|
FP15737
|
FP15737 |
| chr6_-_133084580 | 0.09 |
ENST00000525270.1
ENST00000530536.1 ENST00000524919.1 |
VNN2
|
vanin 2 |
| chr4_-_7873713 | 0.08 |
ENST00000382543.3
|
AFAP1
|
actin filament associated protein 1 |
| chr5_+_35852797 | 0.08 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr16_+_67034466 | 0.08 |
ENST00000535696.1
|
CES4A
|
carboxylesterase 4A |
| chr16_+_67034456 | 0.08 |
ENST00000540579.1
|
CES4A
|
carboxylesterase 4A |
| chr14_-_58894332 | 0.08 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr8_-_81083341 | 0.08 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr4_-_819901 | 0.08 |
ENST00000304062.6
|
CPLX1
|
complexin 1 |
| chr16_+_29674540 | 0.08 |
ENST00000436527.1
ENST00000360121.3 ENST00000449759.1 |
SPN
QPRT
|
sialophorin quinolinate phosphoribosyltransferase |
| chr17_+_7591639 | 0.07 |
ENST00000396463.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr12_-_10320190 | 0.07 |
ENST00000543993.1
ENST00000339968.6 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr7_-_127672146 | 0.07 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr14_-_58894223 | 0.07 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr11_+_18417948 | 0.07 |
ENST00000542179.1
|
LDHA
|
lactate dehydrogenase A |
| chr4_-_100485183 | 0.07 |
ENST00000394876.2
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr10_-_14596140 | 0.07 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr1_+_151512775 | 0.07 |
ENST00000368849.3
ENST00000392712.3 ENST00000353024.3 ENST00000368848.2 ENST00000538902.1 |
TUFT1
|
tuftelin 1 |
| chr12_+_60058458 | 0.07 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_+_55464600 | 0.06 |
ENST00000371265.4
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr3_+_111717511 | 0.06 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr6_-_137113604 | 0.06 |
ENST00000359015.4
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr17_-_9808887 | 0.06 |
ENST00000226193.5
|
RCVRN
|
recoverin |
| chr7_+_18330035 | 0.06 |
ENST00000413509.2
|
HDAC9
|
histone deacetylase 9 |
| chr16_-_3767506 | 0.06 |
ENST00000538171.1
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr11_-_72353451 | 0.06 |
ENST00000376450.3
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr2_+_204801471 | 0.06 |
ENST00000316386.6
ENST00000435193.1 |
ICOS
|
inducible T-cell co-stimulator |
| chrM_+_8527 | 0.06 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr11_-_72353494 | 0.06 |
ENST00000544570.1
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr6_+_30844192 | 0.06 |
ENST00000502955.1
ENST00000505066.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr9_+_131873842 | 0.06 |
ENST00000417728.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr7_+_30634297 | 0.06 |
ENST00000389266.3
|
GARS
|
glycyl-tRNA synthetase |
| chr20_+_47835884 | 0.06 |
ENST00000371764.4
|
DDX27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr1_+_41157361 | 0.06 |
ENST00000427410.2
ENST00000447388.3 ENST00000425457.2 ENST00000453631.1 ENST00000456393.2 |
NFYC
|
nuclear transcription factor Y, gamma |
| chr12_-_49504449 | 0.06 |
ENST00000547675.1
|
LMBR1L
|
limb development membrane protein 1-like |
| chr8_-_144679264 | 0.05 |
ENST00000531953.1
ENST00000526133.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr3_-_8811288 | 0.05 |
ENST00000316793.3
ENST00000431493.1 |
OXTR
|
oxytocin receptor |
| chr21_-_35899113 | 0.05 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr19_+_34856141 | 0.05 |
ENST00000586425.1
|
GPI
|
glucose-6-phosphate isomerase |
| chr4_-_153601136 | 0.05 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr1_+_154401791 | 0.05 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr19_-_38720354 | 0.05 |
ENST00000416611.1
|
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chrX_-_63450480 | 0.05 |
ENST00000362002.2
|
ASB12
|
ankyrin repeat and SOCS box containing 12 |
| chr16_-_66952779 | 0.05 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr16_-_30441293 | 0.05 |
ENST00000565758.1
ENST00000567983.1 ENST00000319285.4 |
DCTPP1
|
dCTP pyrophosphatase 1 |
| chr3_+_111717600 | 0.05 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr5_+_176692466 | 0.05 |
ENST00000508029.1
ENST00000503056.1 |
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr2_-_151905288 | 0.05 |
ENST00000409243.1
|
AC023469.1
|
HCG1817310; Uncharacterized protein |
| chr16_-_3767551 | 0.05 |
ENST00000246957.5
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr6_-_32806506 | 0.05 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr18_-_44497308 | 0.05 |
ENST00000585916.1
ENST00000324794.7 ENST00000545673.1 |
PIAS2
|
protein inhibitor of activated STAT, 2 |
| chr19_-_44259053 | 0.05 |
ENST00000601170.1
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr1_+_74701062 | 0.05 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chrX_-_64196307 | 0.05 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr8_+_128426535 | 0.05 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr15_-_77363375 | 0.05 |
ENST00000559494.1
|
TSPAN3
|
tetraspanin 3 |
| chr17_-_62502399 | 0.05 |
ENST00000450599.2
ENST00000585060.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr6_+_34433844 | 0.05 |
ENST00000244458.2
ENST00000374043.2 |
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr8_-_125551278 | 0.04 |
ENST00000519232.1
ENST00000523888.1 ENST00000522810.1 ENST00000519548.1 ENST00000517678.1 ENST00000605953.1 ENST00000276692.6 |
TATDN1
|
TatD DNase domain containing 1 |
| chr20_-_57089934 | 0.04 |
ENST00000439429.1
ENST00000371149.3 |
APCDD1L
|
adenomatosis polyposis coli down-regulated 1-like |
| chr17_-_62502639 | 0.04 |
ENST00000225792.5
ENST00000581697.1 ENST00000584279.1 ENST00000577922.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr2_+_62132800 | 0.04 |
ENST00000538736.1
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr17_+_37784749 | 0.04 |
ENST00000394265.1
ENST00000394267.2 |
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr16_-_66952742 | 0.04 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chrX_-_9754333 | 0.04 |
ENST00000447366.1
|
GPR143
|
G protein-coupled receptor 143 |
| chr11_-_67141640 | 0.04 |
ENST00000533438.1
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr3_-_196242233 | 0.04 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr1_-_217262969 | 0.04 |
ENST00000361525.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr12_+_13349650 | 0.04 |
ENST00000256951.5
ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1
|
epithelial membrane protein 1 |
| chr6_-_32145861 | 0.04 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr2_-_151395525 | 0.04 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr10_-_118032697 | 0.04 |
ENST00000439649.3
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr3_-_116163830 | 0.04 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr15_+_74911430 | 0.04 |
ENST00000562670.1
ENST00000564096.1 |
CLK3
|
CDC-like kinase 3 |
| chr19_+_52074502 | 0.04 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr3_-_58613323 | 0.04 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr3_-_73673991 | 0.04 |
ENST00000308537.4
ENST00000263666.4 |
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr10_+_71078595 | 0.04 |
ENST00000359426.6
|
HK1
|
hexokinase 1 |
| chr1_+_16083098 | 0.04 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr17_-_62503015 | 0.04 |
ENST00000581806.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr17_+_73455788 | 0.04 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chr1_-_94147385 | 0.04 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr19_+_34856078 | 0.04 |
ENST00000589399.1
ENST00000589640.1 ENST00000591204.1 |
GPI
|
glucose-6-phosphate isomerase |
| chr19_-_44259136 | 0.03 |
ENST00000270066.6
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chrX_-_110039286 | 0.03 |
ENST00000434224.1
|
CHRDL1
|
chordin-like 1 |
| chr1_-_217262933 | 0.03 |
ENST00000359162.2
|
ESRRG
|
estrogen-related receptor gamma |
| chr15_-_77363513 | 0.03 |
ENST00000267970.4
|
TSPAN3
|
tetraspanin 3 |
| chr5_-_179107975 | 0.03 |
ENST00000376974.4
|
CBY3
|
chibby homolog 3 (Drosophila) |
| chr8_+_67341239 | 0.03 |
ENST00000320270.2
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr1_+_224301787 | 0.03 |
ENST00000366862.5
ENST00000424254.2 |
FBXO28
|
F-box protein 28 |
| chr4_-_100484825 | 0.03 |
ENST00000273962.3
ENST00000514547.1 ENST00000455368.2 |
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr14_+_76618242 | 0.03 |
ENST00000557542.1
ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L
|
G patch domain containing 2-like |
| chr3_+_173302222 | 0.03 |
ENST00000361589.4
|
NLGN1
|
neuroligin 1 |
| chr1_+_12040238 | 0.03 |
ENST00000444836.1
ENST00000235329.5 |
MFN2
|
mitofusin 2 |
| chr15_+_86686953 | 0.03 |
ENST00000421325.2
|
AGBL1
|
ATP/GTP binding protein-like 1 |
| chr6_+_143772060 | 0.03 |
ENST00000367591.4
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr12_-_49504655 | 0.03 |
ENST00000551782.1
ENST00000267102.8 |
LMBR1L
|
limb development membrane protein 1-like |
| chr17_+_46184911 | 0.03 |
ENST00000580219.1
ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11
|
sorting nexin 11 |
| chrX_-_48776292 | 0.03 |
ENST00000376509.4
|
PIM2
|
pim-2 oncogene |
| chrX_-_110655306 | 0.03 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr2_-_136288740 | 0.03 |
ENST00000264159.6
ENST00000536680.1 |
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr3_-_187455680 | 0.03 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr17_-_46178527 | 0.03 |
ENST00000393408.3
|
CBX1
|
chromobox homolog 1 |
| chr12_-_118406777 | 0.03 |
ENST00000339824.5
|
KSR2
|
kinase suppressor of ras 2 |
| chr2_+_207024306 | 0.03 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr6_+_69942298 | 0.03 |
ENST00000238918.8
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr15_-_77363441 | 0.03 |
ENST00000346495.2
ENST00000424443.3 ENST00000561277.1 |
TSPAN3
|
tetraspanin 3 |
| chr17_+_62503073 | 0.02 |
ENST00000580188.1
ENST00000581056.1 |
CEP95
|
centrosomal protein 95kDa |
| chr12_-_75603202 | 0.02 |
ENST00000393288.2
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr17_-_33390667 | 0.02 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr8_+_27182862 | 0.02 |
ENST00000521164.1
ENST00000346049.5 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr11_-_104827425 | 0.02 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr13_+_46039037 | 0.02 |
ENST00000349995.5
|
COG3
|
component of oligomeric golgi complex 3 |
| chr15_+_40453204 | 0.02 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr17_+_7591747 | 0.02 |
ENST00000534050.1
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr19_-_11347173 | 0.02 |
ENST00000587656.1
|
DOCK6
|
dedicator of cytokinesis 6 |
| chr3_-_53164423 | 0.02 |
ENST00000467048.1
ENST00000394738.3 ENST00000296292.3 |
RFT1
|
RFT1 homolog (S. cerevisiae) |
| chr7_-_92855762 | 0.02 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr1_-_104239302 | 0.02 |
ENST00000446703.1
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr3_-_114343768 | 0.02 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_-_68547019 | 0.02 |
ENST00000409862.1
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr7_-_156803329 | 0.02 |
ENST00000252971.6
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr17_-_79623597 | 0.02 |
ENST00000574024.1
ENST00000331056.5 |
PDE6G
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr1_+_41157421 | 0.02 |
ENST00000372654.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr1_-_155006224 | 0.02 |
ENST00000368424.3
|
DCST2
|
DC-STAMP domain containing 2 |
| chr22_+_21213771 | 0.02 |
ENST00000439214.1
|
SNAP29
|
synaptosomal-associated protein, 29kDa |
| chr12_-_12837423 | 0.02 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr15_-_99057551 | 0.02 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr2_-_207023918 | 0.02 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr1_+_76251912 | 0.02 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr7_+_30811004 | 0.02 |
ENST00000265299.6
|
FAM188B
|
family with sequence similarity 188, member B |
| chr5_+_74011328 | 0.02 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr11_+_64808675 | 0.02 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr6_-_55740352 | 0.02 |
ENST00000370830.3
|
BMP5
|
bone morphogenetic protein 5 |
| chr18_-_56296182 | 0.02 |
ENST00000361673.3
|
ALPK2
|
alpha-kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.1 | 0.5 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 0.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0031049 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.1 | GO:0044035 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.2 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.1 | GO:0060455 | positive regulation of norepinephrine secretion(GO:0010701) negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.0 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.1 | GO:0015966 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.3 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0060180 | female mating behavior(GO:0060180) |
| 0.0 | 0.4 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.0 | 0.3 | GO:0007567 | parturition(GO:0007567) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.0 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.4 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.0 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.0 | GO:0035643 | L-DOPA receptor activity(GO:0035643) L-DOPA binding(GO:0072544) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.4 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |