Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
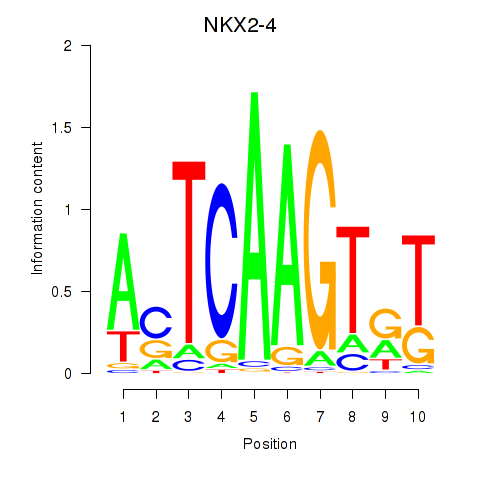
Results for NKX2-4
Z-value: 1.45
Transcription factors associated with NKX2-4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-4
|
ENSG00000125816.4 | NK2 homeobox 4 |
Activity profile of NKX2-4 motif
Sorted Z-values of NKX2-4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_3469181 | 1.02 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr22_-_18923655 | 0.81 |
ENST00000438924.1
ENST00000457083.1 ENST00000420436.1 ENST00000334029.2 ENST00000357068.6 |
PRODH
|
proline dehydrogenase (oxidase) 1 |
| chr6_-_30899924 | 0.76 |
ENST00000359086.3
|
SFTA2
|
surfactant associated 2 |
| chr2_+_109065634 | 0.74 |
ENST00000409821.1
|
GCC2
|
GRIP and coiled-coil domain containing 2 |
| chr1_-_89458287 | 0.67 |
ENST00000370485.2
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chr12_-_58329888 | 0.66 |
ENST00000546580.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr8_+_42396712 | 0.66 |
ENST00000518574.1
ENST00000417410.2 ENST00000414154.2 |
SMIM19
|
small integral membrane protein 19 |
| chr2_-_188419078 | 0.65 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr11_-_111383064 | 0.65 |
ENST00000525791.1
ENST00000456861.2 ENST00000356018.2 |
BTG4
|
B-cell translocation gene 4 |
| chr10_-_28571015 | 0.61 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr19_-_55669093 | 0.58 |
ENST00000344887.5
|
TNNI3
|
troponin I type 3 (cardiac) |
| chr11_+_60699222 | 0.57 |
ENST00000536409.1
|
TMEM132A
|
transmembrane protein 132A |
| chr1_+_144989309 | 0.55 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr1_+_158901329 | 0.54 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr8_+_103876528 | 0.54 |
ENST00000522939.1
ENST00000524007.1 |
KB-1507C5.2
|
HCG15011, isoform CRA_a; Protein LOC100996457 |
| chr7_+_129932974 | 0.53 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr19_-_41942344 | 0.52 |
ENST00000594660.1
|
ATP5SL
|
ATP5S-like |
| chr1_-_100643765 | 0.50 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr11_-_14521349 | 0.49 |
ENST00000534234.1
|
COPB1
|
coatomer protein complex, subunit beta 1 |
| chr19_-_12444519 | 0.48 |
ENST00000595977.1
|
ZNF563
|
zinc finger protein 563 |
| chr6_-_56492816 | 0.48 |
ENST00000522360.1
|
DST
|
dystonin |
| chr9_-_88874519 | 0.48 |
ENST00000376001.3
ENST00000339137.3 |
C9orf153
|
chromosome 9 open reading frame 153 |
| chr4_+_86748898 | 0.47 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr1_-_85040090 | 0.46 |
ENST00000370630.5
|
CTBS
|
chitobiase, di-N-acetyl- |
| chr18_+_9136758 | 0.46 |
ENST00000383440.2
ENST00000262126.4 ENST00000577992.1 |
ANKRD12
|
ankyrin repeat domain 12 |
| chr8_+_67687773 | 0.45 |
ENST00000518388.1
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr5_-_42811986 | 0.45 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr1_+_110036728 | 0.45 |
ENST00000369868.3
ENST00000430195.2 |
CYB561D1
|
cytochrome b561 family, member D1 |
| chr5_-_42812143 | 0.45 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr13_-_41240717 | 0.45 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr8_+_136469684 | 0.44 |
ENST00000355849.5
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3 |
| chr1_-_204183071 | 0.44 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr18_-_60985914 | 0.43 |
ENST00000589955.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr12_-_53601000 | 0.42 |
ENST00000338737.4
ENST00000549086.2 |
ITGB7
|
integrin, beta 7 |
| chr15_+_34394257 | 0.42 |
ENST00000397766.2
|
PGBD4
|
piggyBac transposable element derived 4 |
| chr18_-_28622699 | 0.41 |
ENST00000360428.4
|
DSC3
|
desmocollin 3 |
| chr19_-_49843539 | 0.41 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr17_-_41050716 | 0.41 |
ENST00000417193.1
ENST00000301683.3 ENST00000436546.1 ENST00000431109.2 |
LINC00671
|
long intergenic non-protein coding RNA 671 |
| chr2_-_85828867 | 0.40 |
ENST00000425160.1
|
TMEM150A
|
transmembrane protein 150A |
| chr1_+_61869748 | 0.40 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr18_-_28622774 | 0.39 |
ENST00000434452.1
|
DSC3
|
desmocollin 3 |
| chr11_-_104035088 | 0.39 |
ENST00000302251.5
|
PDGFD
|
platelet derived growth factor D |
| chr4_-_175443943 | 0.39 |
ENST00000296522.6
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr15_+_63354769 | 0.39 |
ENST00000558910.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr2_+_161993465 | 0.38 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr10_+_99332529 | 0.38 |
ENST00000455090.1
|
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr11_+_35965531 | 0.38 |
ENST00000528989.1
ENST00000524419.1 ENST00000315571.5 |
LDLRAD3
|
low density lipoprotein receptor class A domain containing 3 |
| chr15_-_34394008 | 0.36 |
ENST00000527822.1
ENST00000528949.1 |
EMC7
|
ER membrane protein complex subunit 7 |
| chr10_-_18944123 | 0.36 |
ENST00000606425.1
|
RP11-139J15.7
|
Uncharacterized protein |
| chr1_-_197744763 | 0.36 |
ENST00000422998.1
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr1_+_202431859 | 0.35 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr7_-_105221898 | 0.35 |
ENST00000486180.1
ENST00000485614.1 ENST00000480514.1 |
EFCAB10
|
EF-hand calcium binding domain 10 |
| chrX_+_148622138 | 0.34 |
ENST00000450602.2
ENST00000441248.1 |
CXorf40A
|
chromosome X open reading frame 40A |
| chr3_-_107777208 | 0.34 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr4_-_100242549 | 0.34 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr1_+_110036699 | 0.34 |
ENST00000496961.1
ENST00000533024.1 ENST00000310611.4 ENST00000527072.1 ENST00000420578.2 ENST00000528785.1 |
CYB561D1
|
cytochrome b561 family, member D1 |
| chr14_-_59932044 | 0.34 |
ENST00000395116.1
|
GPR135
|
G protein-coupled receptor 135 |
| chr1_+_222886694 | 0.33 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr1_-_70671216 | 0.33 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr4_+_90033968 | 0.32 |
ENST00000317005.2
|
TIGD2
|
tigger transposable element derived 2 |
| chr4_-_75695366 | 0.32 |
ENST00000512743.1
|
BTC
|
betacellulin |
| chr8_+_42396936 | 0.32 |
ENST00000416469.2
|
SMIM19
|
small integral membrane protein 19 |
| chr2_-_188419200 | 0.32 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr21_-_16374688 | 0.31 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr4_-_48683188 | 0.31 |
ENST00000505759.1
|
FRYL
|
FRY-like |
| chr20_+_55108302 | 0.31 |
ENST00000371325.1
|
FAM209B
|
family with sequence similarity 209, member B |
| chr15_-_31523036 | 0.31 |
ENST00000559094.1
ENST00000558388.2 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr17_+_42977122 | 0.31 |
ENST00000412523.2
ENST00000331733.4 ENST00000417826.2 |
FAM187A
CCDC103
|
family with sequence similarity 187, member A coiled-coil domain containing 103 |
| chr7_-_54826869 | 0.31 |
ENST00000450622.1
|
SEC61G
|
Sec61 gamma subunit |
| chr17_-_19648416 | 0.31 |
ENST00000426645.2
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr7_+_36450169 | 0.30 |
ENST00000428612.1
|
ANLN
|
anillin, actin binding protein |
| chr1_-_222886526 | 0.30 |
ENST00000541237.1
|
AIDA
|
axin interactor, dorsalization associated |
| chr4_+_129349188 | 0.29 |
ENST00000511497.1
|
RP11-420A23.1
|
RP11-420A23.1 |
| chr10_-_65225641 | 0.29 |
ENST00000399262.2
|
JMJD1C
|
jumonji domain containing 1C |
| chr15_-_33180439 | 0.29 |
ENST00000559610.1
|
FMN1
|
formin 1 |
| chr1_-_115301235 | 0.29 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr12_+_133757995 | 0.29 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr16_+_8736232 | 0.28 |
ENST00000562973.1
|
METTL22
|
methyltransferase like 22 |
| chr12_+_21590549 | 0.28 |
ENST00000545178.1
ENST00000240651.9 |
PYROXD1
|
pyridine nucleotide-disulphide oxidoreductase domain 1 |
| chr1_-_57431679 | 0.28 |
ENST00000371237.4
ENST00000535057.1 ENST00000543257.1 |
C8B
|
complement component 8, beta polypeptide |
| chr2_-_55920952 | 0.28 |
ENST00000447944.2
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr12_-_53601055 | 0.28 |
ENST00000552972.1
ENST00000422257.3 ENST00000267082.5 |
ITGB7
|
integrin, beta 7 |
| chr6_+_159071015 | 0.28 |
ENST00000360448.3
|
SYTL3
|
synaptotagmin-like 3 |
| chr7_+_65552756 | 0.28 |
ENST00000450043.1
|
AC068533.7
|
AC068533.7 |
| chr8_+_97597148 | 0.28 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr18_+_11851383 | 0.27 |
ENST00000526991.2
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr6_+_7590413 | 0.27 |
ENST00000342415.5
|
SNRNP48
|
small nuclear ribonucleoprotein 48kDa (U11/U12) |
| chr6_-_73935163 | 0.27 |
ENST00000370388.3
|
KHDC1L
|
KH homology domain containing 1-like |
| chr6_-_31088214 | 0.27 |
ENST00000376288.2
|
CDSN
|
corneodesmosin |
| chr2_+_183982255 | 0.27 |
ENST00000455063.1
|
NUP35
|
nucleoporin 35kDa |
| chr10_+_60094735 | 0.27 |
ENST00000373910.4
|
UBE2D1
|
ubiquitin-conjugating enzyme E2D 1 |
| chr12_-_113658892 | 0.27 |
ENST00000299732.2
ENST00000416617.2 |
IQCD
|
IQ motif containing D |
| chr13_-_108870623 | 0.26 |
ENST00000405925.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr7_-_152373216 | 0.26 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr19_-_12444306 | 0.26 |
ENST00000594577.1
ENST00000601858.1 |
ZNF563
|
zinc finger protein 563 |
| chr5_+_154135453 | 0.25 |
ENST00000517616.1
ENST00000518892.1 |
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr10_-_65225722 | 0.25 |
ENST00000399251.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr21_-_15918618 | 0.25 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr18_-_29340827 | 0.25 |
ENST00000269205.5
|
SLC25A52
|
solute carrier family 25, member 52 |
| chr7_+_133812052 | 0.25 |
ENST00000285928.2
|
LRGUK
|
leucine-rich repeats and guanylate kinase domain containing |
| chr11_+_33037652 | 0.25 |
ENST00000311388.3
|
DEPDC7
|
DEP domain containing 7 |
| chr14_-_34420259 | 0.25 |
ENST00000250457.3
ENST00000547327.2 |
EGLN3
|
egl-9 family hypoxia-inducible factor 3 |
| chr10_-_13544945 | 0.24 |
ENST00000378605.3
ENST00000341083.3 |
BEND7
|
BEN domain containing 7 |
| chr5_-_39270725 | 0.24 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr15_-_83474806 | 0.24 |
ENST00000541889.1
ENST00000334574.8 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr12_+_128399965 | 0.24 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chrX_+_105936982 | 0.24 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr13_-_96705624 | 0.24 |
ENST00000376747.3
ENST00000376712.4 ENST00000397618.3 ENST00000376714.3 |
UGGT2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr4_-_68411275 | 0.24 |
ENST00000273853.6
|
CENPC
|
centromere protein C |
| chr4_-_87028478 | 0.24 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr3_-_88108192 | 0.24 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr3_+_177159744 | 0.24 |
ENST00000439009.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr1_-_223308098 | 0.23 |
ENST00000342210.6
|
TLR5
|
toll-like receptor 5 |
| chr2_+_120687335 | 0.23 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr13_-_36944307 | 0.23 |
ENST00000355182.4
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr13_-_108867846 | 0.23 |
ENST00000442234.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr7_+_75932863 | 0.23 |
ENST00000429938.1
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr2_+_10560147 | 0.23 |
ENST00000422133.1
|
HPCAL1
|
hippocalcin-like 1 |
| chr19_-_17559376 | 0.23 |
ENST00000341130.5
|
TMEM221
|
transmembrane protein 221 |
| chr17_+_42923686 | 0.23 |
ENST00000591513.1
|
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr3_+_124303512 | 0.23 |
ENST00000454902.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr14_+_45464658 | 0.23 |
ENST00000555874.1
|
FAM179B
|
family with sequence similarity 179, member B |
| chr5_-_78808617 | 0.23 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr14_+_65381079 | 0.23 |
ENST00000549115.1
ENST00000607599.1 ENST00000548752.2 ENST00000359118.2 ENST00000552002.2 ENST00000551947.1 ENST00000551093.1 ENST00000542227.1 ENST00000447296.2 ENST00000549987.1 |
CHURC1
FNTB
CHURC1-FNTB
|
churchill domain containing 1 farnesyltransferase, CAAX box, beta CHURC1-FNTB readthrough |
| chr5_-_68339648 | 0.23 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr11_-_85780853 | 0.22 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr17_-_5138099 | 0.22 |
ENST00000571800.1
ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr19_-_20748541 | 0.22 |
ENST00000427401.4
ENST00000594419.1 |
ZNF737
|
zinc finger protein 737 |
| chr2_-_37899323 | 0.22 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr1_-_47407136 | 0.22 |
ENST00000462347.1
ENST00000371905.1 ENST00000310638.4 |
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr17_-_38574169 | 0.22 |
ENST00000423485.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr2_-_169887827 | 0.22 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr19_-_20748614 | 0.22 |
ENST00000596797.1
|
ZNF737
|
zinc finger protein 737 |
| chr18_+_21032781 | 0.22 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr10_+_81272287 | 0.22 |
ENST00000520547.2
|
EIF5AL1
|
eukaryotic translation initiation factor 5A-like 1 |
| chr16_-_31100284 | 0.22 |
ENST00000280606.6
|
PRSS53
|
protease, serine, 53 |
| chr14_+_51955831 | 0.22 |
ENST00000356218.4
|
FRMD6
|
FERM domain containing 6 |
| chr12_+_128399917 | 0.21 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr17_+_66244071 | 0.21 |
ENST00000580548.1
ENST00000580753.1 ENST00000392720.2 ENST00000359783.4 ENST00000584837.1 ENST00000579724.1 ENST00000584494.1 ENST00000580837.1 |
AMZ2
|
archaelysin family metallopeptidase 2 |
| chrX_-_54069576 | 0.21 |
ENST00000445025.1
ENST00000322659.8 |
PHF8
|
PHD finger protein 8 |
| chr1_+_95583479 | 0.21 |
ENST00000455656.1
ENST00000604534.1 |
TMEM56
RP11-57H12.6
|
transmembrane protein 56 TMEM56-RWDD3 readthrough |
| chr2_-_225434538 | 0.21 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr9_+_71986182 | 0.21 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chrX_-_44402231 | 0.21 |
ENST00000378045.4
|
FUNDC1
|
FUN14 domain containing 1 |
| chr12_+_133758115 | 0.21 |
ENST00000541009.2
ENST00000592241.1 |
ZNF268
|
zinc finger protein 268 |
| chr22_+_25465786 | 0.21 |
ENST00000401395.1
|
KIAA1671
|
KIAA1671 |
| chr13_-_52026730 | 0.20 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr2_-_202483867 | 0.20 |
ENST00000439802.1
ENST00000286195.3 ENST00000439140.1 ENST00000450242.1 |
ALS2CR11
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 11 |
| chr3_-_126327398 | 0.20 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr18_+_61445205 | 0.20 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr2_+_204571198 | 0.20 |
ENST00000374481.3
ENST00000458610.2 ENST00000324106.8 |
CD28
|
CD28 molecule |
| chr2_-_175462934 | 0.19 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr2_-_179315786 | 0.19 |
ENST00000457633.1
ENST00000438687.3 ENST00000325748.4 |
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr8_-_7320974 | 0.19 |
ENST00000528943.1
ENST00000359758.5 ENST00000361111.2 ENST00000398462.2 ENST00000297498.2 ENST00000317900.5 |
SPAG11B
|
sperm associated antigen 11B |
| chr8_+_7705398 | 0.19 |
ENST00000400125.2
ENST00000434307.2 ENST00000326558.5 ENST00000351436.4 ENST00000528033.1 |
SPAG11A
|
sperm associated antigen 11A |
| chr2_+_191221240 | 0.19 |
ENST00000409027.1
ENST00000458193.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr6_-_134861089 | 0.19 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr8_+_74206829 | 0.19 |
ENST00000240285.5
|
RDH10
|
retinol dehydrogenase 10 (all-trans) |
| chr2_-_25451065 | 0.18 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr2_+_172309634 | 0.18 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr6_-_28303901 | 0.18 |
ENST00000439158.1
ENST00000446474.1 ENST00000414431.1 ENST00000344279.6 ENST00000453745.1 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr5_+_64064748 | 0.18 |
ENST00000381070.3
ENST00000508024.1 |
CWC27
|
CWC27 spliceosome-associated protein homolog (S. cerevisiae) |
| chr4_+_102734967 | 0.18 |
ENST00000444316.2
|
BANK1
|
B-cell scaffold protein with ankyrin repeats 1 |
| chr4_-_103746683 | 0.18 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr15_+_44719996 | 0.18 |
ENST00000559793.1
ENST00000558968.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr19_-_12444491 | 0.18 |
ENST00000293725.5
|
ZNF563
|
zinc finger protein 563 |
| chr4_-_178363581 | 0.18 |
ENST00000264595.2
|
AGA
|
aspartylglucosaminidase |
| chr6_-_143832793 | 0.18 |
ENST00000438118.2
|
FUCA2
|
fucosidase, alpha-L- 2, plasma |
| chr7_-_92463210 | 0.18 |
ENST00000265734.4
|
CDK6
|
cyclin-dependent kinase 6 |
| chr4_-_69111401 | 0.18 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr10_+_22605374 | 0.18 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chr21_+_40823753 | 0.17 |
ENST00000333634.4
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein |
| chr5_-_34919094 | 0.17 |
ENST00000341754.4
|
RAD1
|
RAD1 homolog (S. pombe) |
| chr22_-_26961328 | 0.17 |
ENST00000398110.2
|
TPST2
|
tyrosylprotein sulfotransferase 2 |
| chr7_+_134551583 | 0.17 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr4_+_3768075 | 0.16 |
ENST00000509482.1
ENST00000330055.5 |
ADRA2C
|
adrenoceptor alpha 2C |
| chr6_+_45390222 | 0.16 |
ENST00000359524.5
|
RUNX2
|
runt-related transcription factor 2 |
| chr14_+_62229075 | 0.16 |
ENST00000216294.4
|
SNAPC1
|
small nuclear RNA activating complex, polypeptide 1, 43kDa |
| chr2_+_103378472 | 0.16 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr12_-_102591604 | 0.16 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr14_+_78266408 | 0.16 |
ENST00000238561.5
|
ADCK1
|
aarF domain containing kinase 1 |
| chrX_-_138914394 | 0.16 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr6_-_149969871 | 0.16 |
ENST00000335643.8
ENST00000444282.1 |
KATNA1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr11_+_61717842 | 0.16 |
ENST00000449131.2
|
BEST1
|
bestrophin 1 |
| chr4_-_175041663 | 0.16 |
ENST00000503140.1
|
RP11-148L24.1
|
RP11-148L24.1 |
| chr4_-_103746924 | 0.16 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr11_+_19138670 | 0.16 |
ENST00000446113.2
ENST00000399351.3 |
ZDHHC13
|
zinc finger, DHHC-type containing 13 |
| chr11_-_105892937 | 0.16 |
ENST00000301919.4
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr12_+_18891045 | 0.15 |
ENST00000317658.3
|
CAPZA3
|
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr6_+_153552455 | 0.15 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr14_-_81408093 | 0.15 |
ENST00000555265.1
|
CEP128
|
centrosomal protein 128kDa |
| chr1_+_87794150 | 0.15 |
ENST00000370544.5
|
LMO4
|
LIM domain only 4 |
| chr12_+_123011776 | 0.15 |
ENST00000450485.2
ENST00000333479.7 |
KNTC1
|
kinetochore associated 1 |
| chr1_-_89591749 | 0.15 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr2_+_205410723 | 0.15 |
ENST00000358768.2
ENST00000351153.1 ENST00000349953.3 |
PARD3B
|
par-3 family cell polarity regulator beta |
| chr4_-_168155169 | 0.15 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr10_+_98592674 | 0.15 |
ENST00000356016.3
ENST00000371097.4 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chr14_-_23451467 | 0.15 |
ENST00000555074.1
ENST00000361265.4 |
RP11-298I3.5
AJUBA
|
RP11-298I3.5 ajuba LIM protein |
| chr14_+_56078695 | 0.15 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr19_+_7953417 | 0.15 |
ENST00000600345.1
ENST00000598224.1 |
LRRC8E
|
leucine rich repeat containing 8 family, member E |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 1.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 0.7 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.2 | 0.5 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.2 | 0.5 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 0.7 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.4 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.1 | 0.4 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 0.3 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.3 | GO:0031106 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.1 | 0.3 | GO:2000625 | rRNA import into mitochondrion(GO:0035928) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.1 | 0.3 | GO:0090299 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 | 0.2 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.1 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.4 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.1 | 0.2 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.3 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.1 | 0.4 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.5 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.2 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.2 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.2 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.2 | GO:0032811 | regulation of epinephrine secretion(GO:0014060) negative regulation of epinephrine secretion(GO:0032811) epinephrine secretion(GO:0048242) |
| 0.1 | 0.2 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.1 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.2 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.2 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.0 | 0.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.4 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.3 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.5 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.2 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.3 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.3 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 0.6 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.0 | 0.4 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.3 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.2 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.4 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.3 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.2 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.1 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.0 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.2 | GO:0031274 | regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) |
| 0.0 | 0.3 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.0 | GO:0002892 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) regulation of type II hypersensitivity(GO:0002892) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.0 | 0.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:0002442 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 0.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.5 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.5 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.2 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.5 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.4 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 0.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.2 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.2 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.5 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.8 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.2 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |