Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
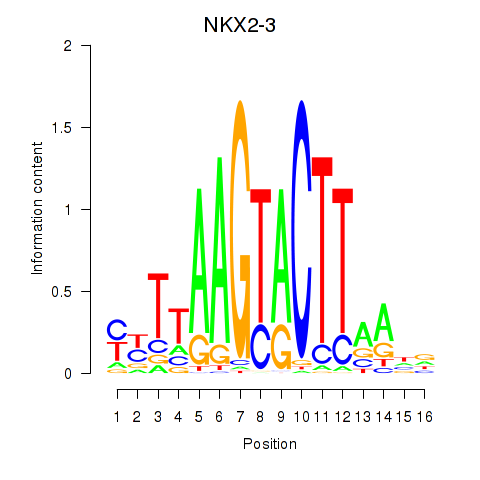
Results for NKX2-3
Z-value: 0.94
Transcription factors associated with NKX2-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-3
|
ENSG00000119919.9 | NK2 homeobox 3 |
Activity profile of NKX2-3 motif
Sorted Z-values of NKX2-3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_114163945 | 0.73 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chrX_+_9502971 | 0.63 |
ENST00000452824.1
|
TBL1X
|
transducin (beta)-like 1X-linked |
| chr13_-_45048386 | 0.61 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr12_+_111051902 | 0.43 |
ENST00000397655.3
ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1
|
tectonic family member 1 |
| chrX_-_108868390 | 0.42 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr3_+_186743261 | 0.42 |
ENST00000423451.1
ENST00000446170.1 |
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr12_-_123565834 | 0.39 |
ENST00000546049.1
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr6_-_10435032 | 0.37 |
ENST00000491317.1
ENST00000496285.1 ENST00000479822.1 ENST00000487130.1 |
LINC00518
|
long intergenic non-protein coding RNA 518 |
| chr12_+_111051832 | 0.35 |
ENST00000550703.2
ENST00000551590.1 |
TCTN1
|
tectonic family member 1 |
| chr2_-_98972468 | 0.35 |
ENST00000454230.1
|
AC092675.3
|
Uncharacterized protein |
| chr14_+_64565442 | 0.35 |
ENST00000553308.1
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr22_+_45714361 | 0.33 |
ENST00000452238.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr6_+_8652370 | 0.33 |
ENST00000503668.1
|
HULC
|
hepatocellular carcinoma up-regulated long non-coding RNA |
| chr2_-_37501692 | 0.33 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr2_+_64073187 | 0.30 |
ENST00000491621.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr1_-_46642154 | 0.29 |
ENST00000540385.1
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr16_-_79804394 | 0.29 |
ENST00000567993.1
|
RP11-345M22.2
|
RP11-345M22.2 |
| chr12_+_25205155 | 0.29 |
ENST00000550945.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr1_-_205391178 | 0.28 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr16_-_18911366 | 0.28 |
ENST00000565224.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr11_-_46638720 | 0.27 |
ENST00000326737.3
|
HARBI1
|
harbinger transposase derived 1 |
| chr1_-_26147149 | 0.27 |
ENST00000536896.1
|
AL020996.1
|
Uncharacterized protein |
| chr7_+_56032652 | 0.27 |
ENST00000437587.1
|
GBAS
|
glioblastoma amplified sequence |
| chr14_+_36295504 | 0.26 |
ENST00000216807.7
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr2_+_204103663 | 0.26 |
ENST00000356079.4
ENST00000429815.2 |
CYP20A1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr5_+_150404904 | 0.26 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr18_-_25616519 | 0.25 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr2_+_71357744 | 0.25 |
ENST00000498451.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr10_+_13203543 | 0.25 |
ENST00000378714.3
ENST00000479669.1 ENST00000484800.2 |
MCM10
|
minichromosome maintenance complex component 10 |
| chr5_-_114631958 | 0.24 |
ENST00000395557.4
|
CCDC112
|
coiled-coil domain containing 112 |
| chr2_-_178128528 | 0.24 |
ENST00000397063.4
ENST00000421929.1 |
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr12_+_122018697 | 0.24 |
ENST00000541574.1
|
RP13-941N14.1
|
RP13-941N14.1 |
| chr18_-_64271316 | 0.24 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr4_+_76439649 | 0.23 |
ENST00000507557.1
|
THAP6
|
THAP domain containing 6 |
| chr3_-_122134882 | 0.22 |
ENST00000330689.4
|
WDR5B
|
WD repeat domain 5B |
| chr12_-_124419531 | 0.21 |
ENST00000514254.2
|
DNAH10OS
|
dynein, axonemal, heavy chain 10 opposite strand |
| chr5_-_159846066 | 0.21 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr5_-_54988448 | 0.20 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr1_+_152943122 | 0.20 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr1_+_205682497 | 0.20 |
ENST00000598338.1
|
AC119673.1
|
AC119673.1 |
| chr2_+_204103733 | 0.20 |
ENST00000443941.1
|
CYP20A1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr5_-_42887494 | 0.19 |
ENST00000514218.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr2_-_188419078 | 0.19 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr14_+_36295638 | 0.19 |
ENST00000543183.1
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr20_-_33539655 | 0.19 |
ENST00000451957.2
|
GSS
|
glutathione synthetase |
| chr8_-_99954788 | 0.19 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr6_-_46138676 | 0.19 |
ENST00000371383.2
ENST00000230565.3 |
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative) |
| chrX_-_16730984 | 0.19 |
ENST00000380241.3
|
CTPS2
|
CTP synthase 2 |
| chr3_+_113775594 | 0.18 |
ENST00000479882.1
ENST00000493014.1 |
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr4_-_104020968 | 0.18 |
ENST00000504285.1
|
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr18_-_51750948 | 0.18 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr19_+_15751689 | 0.18 |
ENST00000586182.2
ENST00000591058.1 ENST00000221307.8 |
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr7_-_29186008 | 0.18 |
ENST00000396276.3
ENST00000265394.5 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr7_-_38948774 | 0.18 |
ENST00000395969.2
ENST00000414632.1 ENST00000310301.4 |
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr10_-_70092671 | 0.18 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr7_+_141463897 | 0.17 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr19_+_18485509 | 0.17 |
ENST00000597765.1
|
GDF15
|
growth differentiation factor 15 |
| chr11_+_65266507 | 0.17 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr12_-_2966193 | 0.16 |
ENST00000382678.3
|
AC005841.1
|
Uncharacterized protein ENSP00000372125 |
| chr6_+_134758827 | 0.16 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr8_-_17555164 | 0.16 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr4_-_75695366 | 0.16 |
ENST00000512743.1
|
BTC
|
betacellulin |
| chr15_+_58702742 | 0.16 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr5_+_150051149 | 0.16 |
ENST00000523553.1
|
MYOZ3
|
myozenin 3 |
| chr14_+_67831576 | 0.16 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr4_+_48343339 | 0.16 |
ENST00000264313.6
|
SLAIN2
|
SLAIN motif family, member 2 |
| chr11_-_65363259 | 0.16 |
ENST00000342202.4
|
KCNK7
|
potassium channel, subfamily K, member 7 |
| chr10_-_76868931 | 0.16 |
ENST00000372700.3
ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13
|
dual specificity phosphatase 13 |
| chr3_-_69062764 | 0.16 |
ENST00000295571.5
|
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr4_-_36246060 | 0.16 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr13_-_50018140 | 0.16 |
ENST00000410043.1
ENST00000347776.5 |
CAB39L
|
calcium binding protein 39-like |
| chr2_+_242312264 | 0.16 |
ENST00000445489.1
|
FARP2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr12_-_42719885 | 0.15 |
ENST00000552673.1
ENST00000266529.3 ENST00000552235.1 |
ZCRB1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr10_-_73976884 | 0.15 |
ENST00000317126.4
ENST00000545550.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr12_-_120315074 | 0.15 |
ENST00000261833.7
ENST00000392521.2 |
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr3_-_101039402 | 0.15 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr14_-_69263043 | 0.15 |
ENST00000408913.2
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr12_+_41136144 | 0.15 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr10_+_35464513 | 0.15 |
ENST00000494479.1
ENST00000463314.1 ENST00000342105.3 ENST00000495301.1 ENST00000463960.1 |
CREM
|
cAMP responsive element modulator |
| chrX_-_134049262 | 0.15 |
ENST00000370783.3
|
MOSPD1
|
motile sperm domain containing 1 |
| chr2_-_188419200 | 0.15 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr7_-_76829125 | 0.15 |
ENST00000248598.5
|
FGL2
|
fibrinogen-like 2 |
| chr3_-_123512688 | 0.14 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chrX_+_37865804 | 0.14 |
ENST00000297875.2
ENST00000357972.5 |
SYTL5
|
synaptotagmin-like 5 |
| chr2_+_64069240 | 0.14 |
ENST00000497883.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr4_-_100242549 | 0.14 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr6_-_32731243 | 0.14 |
ENST00000427449.1
ENST00000411527.1 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr5_-_180688105 | 0.14 |
ENST00000327767.4
|
TRIM52
|
tripartite motif containing 52 |
| chr12_-_42878101 | 0.14 |
ENST00000552108.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr12_-_104531945 | 0.14 |
ENST00000551446.1
|
NFYB
|
nuclear transcription factor Y, beta |
| chr3_-_131753830 | 0.14 |
ENST00000429747.1
|
CPNE4
|
copine IV |
| chr9_-_75488984 | 0.14 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr4_-_52883786 | 0.14 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr22_+_45714672 | 0.13 |
ENST00000424557.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr4_-_90758227 | 0.13 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chrX_-_108976410 | 0.13 |
ENST00000504980.1
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4 |
| chr1_+_174669653 | 0.13 |
ENST00000325589.5
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr5_-_132200477 | 0.13 |
ENST00000296875.2
|
GDF9
|
growth differentiation factor 9 |
| chr1_+_174670143 | 0.13 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr11_-_71752838 | 0.13 |
ENST00000537930.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr18_+_43684298 | 0.13 |
ENST00000282058.6
|
HAUS1
|
HAUS augmin-like complex, subunit 1 |
| chr12_+_42719947 | 0.13 |
ENST00000395580.3
ENST00000337898.6 ENST00000358314.7 ENST00000395568.2 ENST00000256678.8 ENST00000449194.2 ENST00000552761.1 ENST00000317560.9 ENST00000432191.2 |
PPHLN1
|
periphilin 1 |
| chr4_-_90758118 | 0.13 |
ENST00000420646.2
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr1_+_43824669 | 0.13 |
ENST00000372462.1
|
CDC20
|
cell division cycle 20 |
| chr14_+_55494323 | 0.13 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr12_-_102591604 | 0.13 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr3_+_25831567 | 0.13 |
ENST00000280701.3
ENST00000420173.2 |
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr5_-_110848189 | 0.13 |
ENST00000296632.3
ENST00000512160.1 ENST00000509887.1 |
STARD4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr15_+_69452959 | 0.12 |
ENST00000261858.2
|
GLCE
|
glucuronic acid epimerase |
| chr14_-_50154921 | 0.12 |
ENST00000553805.2
ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2
|
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr5_+_112312399 | 0.12 |
ENST00000515408.1
ENST00000513585.1 |
DCP2
|
decapping mRNA 2 |
| chr3_+_145782358 | 0.12 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chr2_-_36825281 | 0.12 |
ENST00000405912.3
ENST00000379245.4 |
FEZ2
|
fasciculation and elongation protein zeta 2 (zygin II) |
| chr9_-_138391692 | 0.12 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr4_+_189321881 | 0.12 |
ENST00000512839.1
ENST00000513313.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
| chrX_+_69509870 | 0.12 |
ENST00000374388.3
|
KIF4A
|
kinesin family member 4A |
| chr14_+_100531738 | 0.12 |
ENST00000555706.1
|
EVL
|
Enah/Vasp-like |
| chr9_+_131447342 | 0.12 |
ENST00000409104.3
|
SET
|
SET nuclear oncogene |
| chr14_-_90798273 | 0.12 |
ENST00000357904.3
|
NRDE2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr16_+_56969284 | 0.12 |
ENST00000568358.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr2_+_202125219 | 0.12 |
ENST00000323492.7
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr19_-_1401486 | 0.12 |
ENST00000252288.2
ENST00000447102.3 |
GAMT
|
guanidinoacetate N-methyltransferase |
| chr3_+_182971018 | 0.12 |
ENST00000326505.3
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr7_+_6071007 | 0.12 |
ENST00000409061.1
|
ANKRD61
|
ankyrin repeat domain 61 |
| chr4_+_139230865 | 0.12 |
ENST00000502757.1
ENST00000507145.1 ENST00000515282.1 ENST00000510736.1 |
LINC00499
|
long intergenic non-protein coding RNA 499 |
| chr17_+_61905058 | 0.11 |
ENST00000375812.4
ENST00000581882.1 |
PSMC5
|
proteasome (prosome, macropain) 26S subunit, ATPase, 5 |
| chr1_+_152730499 | 0.11 |
ENST00000368773.1
|
KPRP
|
keratinocyte proline-rich protein |
| chrX_-_106362013 | 0.11 |
ENST00000372487.1
ENST00000372479.3 ENST00000203616.8 |
RBM41
|
RNA binding motif protein 41 |
| chr3_+_23847432 | 0.11 |
ENST00000346855.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr22_-_21213676 | 0.11 |
ENST00000449120.1
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha |
| chr17_+_61904766 | 0.11 |
ENST00000581842.1
ENST00000582130.1 ENST00000584320.1 ENST00000585123.1 ENST00000580864.1 |
PSMC5
|
proteasome (prosome, macropain) 26S subunit, ATPase, 5 |
| chr7_-_86849836 | 0.11 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr12_+_116985896 | 0.11 |
ENST00000547114.1
|
RP11-809C9.2
|
RP11-809C9.2 |
| chr4_-_39034542 | 0.11 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr2_+_64069459 | 0.11 |
ENST00000445915.2
ENST00000475462.1 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr22_-_30814469 | 0.11 |
ENST00000598426.1
|
KIAA1658
|
KIAA1658 |
| chr3_-_180397256 | 0.11 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr4_-_120550146 | 0.11 |
ENST00000354960.3
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr7_-_130353553 | 0.11 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr5_-_142814241 | 0.10 |
ENST00000504572.1
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr5_+_138210919 | 0.10 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr4_-_39033963 | 0.10 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr3_+_113775576 | 0.10 |
ENST00000485050.1
ENST00000281273.4 |
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr13_-_50018241 | 0.10 |
ENST00000409308.1
|
CAB39L
|
calcium binding protein 39-like |
| chr2_+_64068844 | 0.10 |
ENST00000337130.5
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr3_+_57094469 | 0.10 |
ENST00000334325.1
|
SPATA12
|
spermatogenesis associated 12 |
| chr11_-_75017734 | 0.10 |
ENST00000532525.1
|
ARRB1
|
arrestin, beta 1 |
| chr10_-_27529486 | 0.10 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chrX_-_21776281 | 0.10 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr12_+_19389814 | 0.10 |
ENST00000536974.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr1_-_212965104 | 0.10 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr11_+_17599811 | 0.10 |
ENST00000342528.2
|
OTOG
|
otogelin |
| chr16_-_66952779 | 0.10 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr5_-_110848253 | 0.10 |
ENST00000505803.1
ENST00000502322.1 |
STARD4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr16_+_23765948 | 0.10 |
ENST00000300113.2
|
CHP2
|
calcineurin-like EF-hand protein 2 |
| chr5_-_96518907 | 0.09 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
| chr12_+_57390745 | 0.09 |
ENST00000600202.1
|
HBCBP
|
HBcAg-binding protein; Uncharacterized protein |
| chr12_+_6309963 | 0.09 |
ENST00000382515.2
|
CD9
|
CD9 molecule |
| chr15_-_90437195 | 0.09 |
ENST00000560940.1
ENST00000558011.1 |
AP3S2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chrX_-_49121165 | 0.09 |
ENST00000376207.4
ENST00000376199.2 |
FOXP3
|
forkhead box P3 |
| chr4_+_76439665 | 0.09 |
ENST00000508105.1
ENST00000311638.3 ENST00000380837.3 ENST00000507556.1 ENST00000504190.1 ENST00000507885.1 ENST00000502620.1 ENST00000514480.1 |
THAP6
|
THAP domain containing 6 |
| chr3_-_122233723 | 0.09 |
ENST00000493510.1
ENST00000344337.6 ENST00000476916.1 ENST00000465882.1 |
KPNA1
|
karyopherin alpha 1 (importin alpha 5) |
| chr18_+_32173276 | 0.09 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr7_+_1609765 | 0.09 |
ENST00000437964.1
ENST00000533935.1 ENST00000532358.1 ENST00000524978.1 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr7_-_80548667 | 0.09 |
ENST00000265361.3
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr3_-_42623805 | 0.09 |
ENST00000456515.1
|
SEC22C
|
SEC22 vesicle trafficking protein homolog C (S. cerevisiae) |
| chr8_-_101718991 | 0.09 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr7_-_2883650 | 0.09 |
ENST00000544127.1
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr15_-_101142362 | 0.09 |
ENST00000559577.1
ENST00000561308.1 ENST00000560133.1 ENST00000560941.1 ENST00000559736.1 ENST00000560272.1 |
LINS
|
lines homolog (Drosophila) |
| chr2_-_172291273 | 0.09 |
ENST00000442778.1
ENST00000453846.1 |
METTL8
|
methyltransferase like 8 |
| chrX_-_134049233 | 0.09 |
ENST00000370779.4
|
MOSPD1
|
motile sperm domain containing 1 |
| chr6_-_150212029 | 0.09 |
ENST00000529948.1
ENST00000357183.4 ENST00000367363.3 |
RAET1E
|
retinoic acid early transcript 1E |
| chr19_+_50148087 | 0.09 |
ENST00000601038.1
ENST00000595242.1 |
SCAF1
|
SR-related CTD-associated factor 1 |
| chr12_-_53189892 | 0.09 |
ENST00000309505.3
ENST00000417996.2 |
KRT3
|
keratin 3 |
| chr6_+_31126291 | 0.09 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr19_-_46288917 | 0.09 |
ENST00000537879.1
ENST00000596586.1 ENST00000595946.1 |
DMWD
AC011530.4
|
dystrophia myotonica, WD repeat containing Uncharacterized protein |
| chr1_-_89488510 | 0.09 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr1_+_202317855 | 0.09 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr12_+_10460417 | 0.09 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr1_+_65613340 | 0.09 |
ENST00000546702.1
|
AK4
|
adenylate kinase 4 |
| chr16_+_68877496 | 0.08 |
ENST00000261778.1
|
TANGO6
|
transport and golgi organization 6 homolog (Drosophila) |
| chr19_+_42746927 | 0.08 |
ENST00000378108.1
|
AC006486.1
|
AC006486.1 |
| chr12_-_122711968 | 0.08 |
ENST00000485724.1
|
DIABLO
|
diablo, IAP-binding mitochondrial protein |
| chr1_-_33116128 | 0.08 |
ENST00000436661.1
ENST00000373501.2 ENST00000341885.5 ENST00000468695.1 |
ZBTB8OS
|
zinc finger and BTB domain containing 8 opposite strand |
| chr1_+_65613217 | 0.08 |
ENST00000545314.1
|
AK4
|
adenylate kinase 4 |
| chr12_+_12966250 | 0.08 |
ENST00000352940.4
ENST00000358007.3 ENST00000544400.1 |
DDX47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr6_-_111927062 | 0.08 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr11_+_118889142 | 0.08 |
ENST00000533632.1
|
TRAPPC4
|
trafficking protein particle complex 4 |
| chr11_+_12399071 | 0.08 |
ENST00000539723.1
ENST00000550549.1 |
PARVA
|
parvin, alpha |
| chr20_-_31071309 | 0.08 |
ENST00000326071.4
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr4_-_104021009 | 0.08 |
ENST00000509245.1
ENST00000296424.4 |
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr18_+_43684310 | 0.08 |
ENST00000592471.1
ENST00000585518.1 |
HAUS1
|
HAUS augmin-like complex, subunit 1 |
| chr3_-_49058479 | 0.08 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr6_+_134273300 | 0.08 |
ENST00000416965.1
|
TBPL1
|
TBP-like 1 |
| chr2_-_231989808 | 0.08 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr1_+_206317450 | 0.08 |
ENST00000358184.2
ENST00000361052.3 ENST00000360218.2 |
CTSE
|
cathepsin E |
| chr7_-_86849883 | 0.08 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr9_-_39239171 | 0.08 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr5_-_39270725 | 0.08 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr5_-_83680192 | 0.07 |
ENST00000380138.3
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.1 | 0.3 | GO:0010877 | lipid transport involved in lipid storage(GO:0010877) |
| 0.1 | 0.7 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.3 | GO:2000470 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.2 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.3 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.2 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0036101 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.2 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.1 | GO:2000532 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) regulation of tolerance induction dependent upon immune response(GO:0002652) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.1 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.3 | GO:2000809 | regulation of postsynaptic density protein 95 clustering(GO:1902897) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:0039650 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.2 | GO:1902846 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.5 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.2 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.2 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.0 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.2 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.2 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |