Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
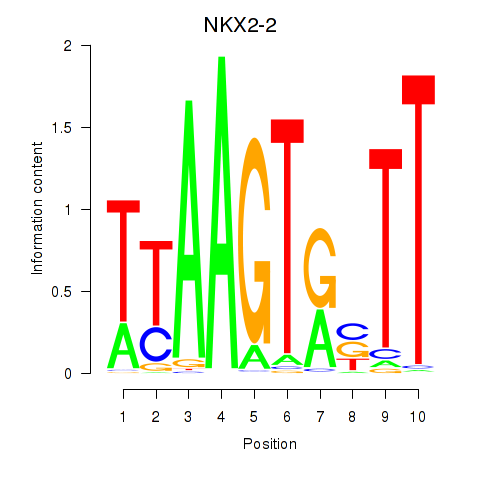
Results for NKX2-2
Z-value: 0.54
Transcription factors associated with NKX2-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-2
|
ENSG00000125820.5 | NK2 homeobox 2 |
Activity profile of NKX2-2 motif
Sorted Z-values of NKX2-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_37723336 | 1.42 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr12_+_78359999 | 0.73 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr17_+_38171681 | 0.69 |
ENST00000225474.2
ENST00000331769.2 ENST00000394148.3 ENST00000577675.1 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr17_-_38859996 | 0.60 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr17_+_38171614 | 0.49 |
ENST00000583218.1
ENST00000394149.3 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr1_-_153363452 | 0.44 |
ENST00000368732.1
ENST00000368733.3 |
S100A8
|
S100 calcium binding protein A8 |
| chrX_+_115567767 | 0.41 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr3_-_152058779 | 0.34 |
ENST00000408960.3
|
TMEM14E
|
transmembrane protein 14E |
| chr7_+_37723450 | 0.28 |
ENST00000447769.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr12_-_49582978 | 0.26 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr3_+_142838091 | 0.22 |
ENST00000309575.3
|
CHST2
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
| chr2_-_36779411 | 0.20 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr19_-_44174330 | 0.19 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr2_+_152214098 | 0.19 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr1_+_32666188 | 0.18 |
ENST00000421922.2
|
CCDC28B
|
coiled-coil domain containing 28B |
| chr5_+_147582348 | 0.18 |
ENST00000514389.1
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr4_+_89299885 | 0.17 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr14_+_23654525 | 0.17 |
ENST00000399910.1
ENST00000492621.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chr4_-_76957214 | 0.17 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr3_-_121379739 | 0.16 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr19_-_44174305 | 0.16 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr14_+_21467414 | 0.16 |
ENST00000554422.1
ENST00000298681.4 |
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr19_-_51568324 | 0.15 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr3_+_98482175 | 0.15 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr6_-_72129806 | 0.15 |
ENST00000413945.1
ENST00000602878.1 ENST00000436803.1 ENST00000421704.1 ENST00000441570.1 |
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr21_+_30968360 | 0.15 |
ENST00000333765.4
|
GRIK1-AS2
|
GRIK1 antisense RNA 2 |
| chr5_+_167718604 | 0.15 |
ENST00000265293.4
|
WWC1
|
WW and C2 domain containing 1 |
| chr1_-_26633480 | 0.14 |
ENST00000450041.1
|
UBXN11
|
UBX domain protein 11 |
| chr5_+_148206156 | 0.14 |
ENST00000305988.4
|
ADRB2
|
adrenoceptor beta 2, surface |
| chr19_-_17366257 | 0.14 |
ENST00000594059.1
|
AC010646.3
|
Uncharacterized protein |
| chr3_-_114790179 | 0.14 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_-_70409953 | 0.14 |
ENST00000419381.1
|
C2orf42
|
chromosome 2 open reading frame 42 |
| chr8_-_95220775 | 0.13 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr5_-_66942617 | 0.13 |
ENST00000507298.1
|
RP11-83M16.5
|
RP11-83M16.5 |
| chr8_-_133772870 | 0.12 |
ENST00000522334.1
ENST00000519016.1 |
TMEM71
|
transmembrane protein 71 |
| chr9_+_99690592 | 0.12 |
ENST00000354649.3
|
NUTM2G
|
NUT family member 2G |
| chr11_-_7698453 | 0.12 |
ENST00000524608.1
|
CYB5R2
|
cytochrome b5 reductase 2 |
| chr17_+_61086917 | 0.12 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr5_+_159656437 | 0.12 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr14_+_39703112 | 0.11 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chrY_+_15418467 | 0.11 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr2_-_214015111 | 0.11 |
ENST00000433134.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr1_+_15272271 | 0.11 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr17_+_18647326 | 0.11 |
ENST00000395667.1
ENST00000395665.4 ENST00000308799.4 ENST00000301938.4 |
FBXW10
|
F-box and WD repeat domain containing 10 |
| chr12_+_76653682 | 0.11 |
ENST00000553247.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chr15_-_60683326 | 0.11 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr22_-_42086477 | 0.10 |
ENST00000402458.1
|
NHP2L1
|
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chr19_-_39881669 | 0.10 |
ENST00000221266.7
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chrX_-_19817869 | 0.10 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr6_+_151561085 | 0.10 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr11_+_93479588 | 0.10 |
ENST00000526335.1
|
C11orf54
|
chromosome 11 open reading frame 54 |
| chr18_-_68004529 | 0.10 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chrX_-_130037162 | 0.10 |
ENST00000432489.1
|
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr4_-_178169927 | 0.09 |
ENST00000512516.1
|
RP11-487E13.1
|
Uncharacterized protein |
| chr8_+_11666649 | 0.09 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr8_-_133772794 | 0.09 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr10_+_32735030 | 0.09 |
ENST00000277657.6
ENST00000362006.5 |
CCDC7
|
coiled-coil domain containing 7 |
| chr5_-_78281603 | 0.09 |
ENST00000264914.4
|
ARSB
|
arylsulfatase B |
| chr10_-_36812323 | 0.09 |
ENST00000543053.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr11_-_26743546 | 0.09 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr8_+_11660227 | 0.09 |
ENST00000443614.2
ENST00000525900.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr9_-_85882145 | 0.09 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr17_+_7591639 | 0.09 |
ENST00000396463.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr4_-_54518619 | 0.09 |
ENST00000507168.1
ENST00000510143.1 |
LNX1
|
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| chr10_-_112678692 | 0.09 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr2_-_203735484 | 0.09 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr3_-_57326704 | 0.08 |
ENST00000487349.1
ENST00000389601.3 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr14_-_38036271 | 0.08 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr4_-_105416039 | 0.08 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr2_-_151905288 | 0.08 |
ENST00000409243.1
|
AC023469.1
|
HCG1817310; Uncharacterized protein |
| chr3_+_108541545 | 0.08 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr14_+_94547675 | 0.08 |
ENST00000393115.3
ENST00000554166.1 ENST00000556381.1 ENST00000553664.1 ENST00000555341.1 ENST00000557218.1 ENST00000554544.1 ENST00000557066.1 |
IFI27L1
|
interferon, alpha-inducible protein 27-like 1 |
| chr10_-_121296045 | 0.08 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr2_+_219110149 | 0.08 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr19_-_18433910 | 0.08 |
ENST00000594828.3
ENST00000593829.1 |
LSM4
|
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr1_+_48569306 | 0.08 |
ENST00000601108.1
|
AL109659.1
|
HCG1780467; PRO0529; Uncharacterized protein |
| chr14_-_38028689 | 0.08 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr7_-_7782204 | 0.08 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr3_+_167582561 | 0.08 |
ENST00000463642.1
ENST00000464514.1 |
RP11-298O21.6
RP11-298O21.7
|
RP11-298O21.6 RP11-298O21.7 |
| chr7_+_2394445 | 0.08 |
ENST00000360876.4
ENST00000413917.1 ENST00000397011.2 |
EIF3B
|
eukaryotic translation initiation factor 3, subunit B |
| chr20_+_30102231 | 0.07 |
ENST00000335574.5
ENST00000340852.5 ENST00000398174.3 ENST00000376127.3 ENST00000344042.5 |
HM13
|
histocompatibility (minor) 13 |
| chr18_+_55888767 | 0.07 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr10_-_36813162 | 0.07 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr3_+_101280677 | 0.07 |
ENST00000309922.6
ENST00000495642.1 |
TRMT10C
|
tRNA methyltransferase 10 homolog C (S. cerevisiae) |
| chr19_+_39616410 | 0.07 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr16_-_1464688 | 0.07 |
ENST00000389221.4
ENST00000508903.2 ENST00000397462.1 ENST00000301712.5 |
UNKL
|
unkempt family zinc finger-like |
| chr22_-_43567750 | 0.07 |
ENST00000494035.1
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12 |
| chrX_-_153363125 | 0.07 |
ENST00000407218.1
ENST00000453960.2 |
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chrX_-_130037198 | 0.07 |
ENST00000370935.1
ENST00000338144.3 ENST00000394363.1 |
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr15_-_60690163 | 0.07 |
ENST00000558998.1
ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2
|
annexin A2 |
| chr7_-_944631 | 0.07 |
ENST00000453175.2
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr17_-_29624343 | 0.07 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr18_-_70931689 | 0.07 |
ENST00000581862.1
|
RP11-169F17.1
|
Protein LOC400655 |
| chr12_-_28124903 | 0.07 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr5_+_35852797 | 0.07 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr19_-_3971050 | 0.07 |
ENST00000545797.2
ENST00000596311.1 |
DAPK3
|
death-associated protein kinase 3 |
| chr8_+_11660120 | 0.07 |
ENST00000220584.4
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr3_+_75713481 | 0.07 |
ENST00000308062.3
ENST00000464571.1 |
FRG2C
|
FSHD region gene 2 family, member C |
| chr1_-_161046266 | 0.07 |
ENST00000453926.2
|
PVRL4
|
poliovirus receptor-related 4 |
| chrX_+_105066524 | 0.07 |
ENST00000243300.9
ENST00000428173.2 |
NRK
|
Nik related kinase |
| chr4_+_80584903 | 0.06 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr6_-_116833500 | 0.06 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr6_+_77484663 | 0.06 |
ENST00000607287.1
|
RP11-354K4.2
|
RP11-354K4.2 |
| chr1_-_159880159 | 0.06 |
ENST00000599780.1
|
AL590560.1
|
HCG1995379; Uncharacterized protein |
| chr5_+_140552218 | 0.06 |
ENST00000231137.3
|
PCDHB7
|
protocadherin beta 7 |
| chr1_-_198906528 | 0.06 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr11_-_64900791 | 0.06 |
ENST00000531018.1
|
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr1_-_45253377 | 0.06 |
ENST00000372207.3
|
BEST4
|
bestrophin 4 |
| chr19_-_14889349 | 0.06 |
ENST00000315576.3
ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr4_-_40477766 | 0.06 |
ENST00000507180.1
|
RBM47
|
RNA binding motif protein 47 |
| chr17_+_7591747 | 0.06 |
ENST00000534050.1
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr8_-_99955042 | 0.06 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr5_-_150521192 | 0.06 |
ENST00000523714.1
ENST00000521749.1 |
ANXA6
|
annexin A6 |
| chr6_-_113754604 | 0.06 |
ENST00000421737.1
|
RP1-124C6.1
|
RP1-124C6.1 |
| chr14_-_24020858 | 0.06 |
ENST00000419474.3
|
ZFHX2
|
zinc finger homeobox 2 |
| chr13_-_20357057 | 0.06 |
ENST00000338910.4
|
PSPC1
|
paraspeckle component 1 |
| chr14_+_94547628 | 0.06 |
ENST00000555523.1
|
IFI27L1
|
interferon, alpha-inducible protein 27-like 1 |
| chr5_+_147582387 | 0.06 |
ENST00000325630.2
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr3_-_33686743 | 0.05 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr13_+_111855399 | 0.05 |
ENST00000426768.2
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr10_+_90346519 | 0.05 |
ENST00000371939.3
|
LIPJ
|
lipase, family member J |
| chr18_+_32558380 | 0.05 |
ENST00000588349.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr1_-_41328018 | 0.05 |
ENST00000372638.2
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr21_-_19858196 | 0.05 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr12_+_76653611 | 0.05 |
ENST00000550380.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chr4_+_95376396 | 0.05 |
ENST00000508216.1
ENST00000514743.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr16_+_23194033 | 0.05 |
ENST00000300061.2
|
SCNN1G
|
sodium channel, non-voltage-gated 1, gamma subunit |
| chr7_+_16700806 | 0.05 |
ENST00000446596.1
ENST00000438834.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr19_+_49838653 | 0.05 |
ENST00000598095.1
ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37
|
CD37 molecule |
| chr1_-_213188772 | 0.05 |
ENST00000544555.1
|
ANGEL2
|
angel homolog 2 (Drosophila) |
| chr13_+_25875662 | 0.05 |
ENST00000381736.3
ENST00000463407.1 ENST00000381718.3 |
NUPL1
|
nucleoporin like 1 |
| chr10_+_81462983 | 0.05 |
ENST00000448135.1
ENST00000429828.1 ENST00000372321.1 |
NUTM2B
|
NUT family member 2B |
| chr9_+_103340354 | 0.05 |
ENST00000307584.5
|
MURC
|
muscle-related coiled-coil protein |
| chr15_+_56657613 | 0.05 |
ENST00000352903.2
ENST00000537232.1 ENST00000561221.2 ENST00000558083.2 |
TEX9
|
testis expressed 9 |
| chr6_+_151561506 | 0.05 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr16_-_56223480 | 0.05 |
ENST00000565155.1
|
RP11-461O7.1
|
RP11-461O7.1 |
| chr18_+_3466248 | 0.05 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr11_+_34654011 | 0.05 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr4_-_71532207 | 0.05 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr5_+_154181816 | 0.05 |
ENST00000518677.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr1_+_84630352 | 0.05 |
ENST00000450730.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr3_-_33686925 | 0.05 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr17_+_15604513 | 0.05 |
ENST00000481540.1
|
ZNF286A
|
Homo sapiens zinc finger protein 286A (ZNF286A), transcript variant 6, mRNA. |
| chr4_+_3076388 | 0.05 |
ENST00000355072.5
|
HTT
|
huntingtin |
| chr13_+_25875785 | 0.05 |
ENST00000381747.3
|
NUPL1
|
nucleoporin like 1 |
| chr2_-_151344172 | 0.05 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr1_+_50459990 | 0.05 |
ENST00000448346.1
|
AL645730.2
|
AL645730.2 |
| chr2_+_162272605 | 0.05 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr18_-_43684230 | 0.05 |
ENST00000592989.1
ENST00000589869.1 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr18_+_72201829 | 0.04 |
ENST00000582365.1
|
CNDP1
|
carnosine dipeptidase 1 (metallopeptidase M20 family) |
| chr7_+_80253387 | 0.04 |
ENST00000438020.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr12_+_116955659 | 0.04 |
ENST00000552992.1
|
RP11-148B3.1
|
RP11-148B3.1 |
| chr1_+_61547894 | 0.04 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr2_+_103035102 | 0.04 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr4_-_119759795 | 0.04 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr4_+_139230865 | 0.04 |
ENST00000502757.1
ENST00000507145.1 ENST00000515282.1 ENST00000510736.1 |
LINC00499
|
long intergenic non-protein coding RNA 499 |
| chr9_-_98079965 | 0.04 |
ENST00000289081.3
|
FANCC
|
Fanconi anemia, complementation group C |
| chr8_-_102181718 | 0.04 |
ENST00000565617.1
|
KB-1460A1.5
|
KB-1460A1.5 |
| chr2_+_102953608 | 0.04 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr21_+_17214724 | 0.04 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr19_+_47634039 | 0.04 |
ENST00000597808.1
ENST00000413379.3 ENST00000600706.1 ENST00000540850.1 ENST00000598840.1 ENST00000600753.1 ENST00000270225.7 ENST00000392776.3 |
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr9_-_10612703 | 0.04 |
ENST00000463477.1
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D |
| chr10_-_61513146 | 0.04 |
ENST00000430431.1
|
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr14_+_32963433 | 0.04 |
ENST00000554410.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr7_+_18548878 | 0.04 |
ENST00000456174.2
|
HDAC9
|
histone deacetylase 9 |
| chr2_+_173724771 | 0.04 |
ENST00000538974.1
ENST00000540783.1 |
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr16_+_30212050 | 0.04 |
ENST00000563322.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr10_-_104262426 | 0.04 |
ENST00000487599.1
|
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr2_-_160472052 | 0.04 |
ENST00000437839.1
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr10_+_81601114 | 0.04 |
ENST00000602967.1
ENST00000429984.3 |
NUTM2E
|
NUT family member 2E |
| chr4_-_140477928 | 0.04 |
ENST00000274031.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr1_-_98515395 | 0.04 |
ENST00000424528.2
|
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr2_+_113299990 | 0.04 |
ENST00000537335.1
ENST00000417433.2 |
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr4_+_74269956 | 0.04 |
ENST00000295897.4
ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB
|
albumin |
| chr5_-_139937895 | 0.04 |
ENST00000336283.6
|
SRA1
|
steroid receptor RNA activator 1 |
| chr10_-_104262460 | 0.04 |
ENST00000446605.2
ENST00000369905.4 ENST00000545684.1 |
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr15_-_56209306 | 0.04 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr7_+_2393714 | 0.04 |
ENST00000431643.1
|
EIF3B
|
eukaryotic translation initiation factor 3, subunit B |
| chr11_-_8892900 | 0.04 |
ENST00000526155.1
ENST00000524757.1 ENST00000527392.1 ENST00000534665.1 ENST00000525169.1 ENST00000527516.1 ENST00000533471.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr3_+_189507523 | 0.04 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr16_+_10837643 | 0.04 |
ENST00000574334.1
ENST00000283027.5 ENST00000433392.2 |
NUBP1
|
nucleotide binding protein 1 |
| chr1_-_202936394 | 0.04 |
ENST00000367249.4
|
CYB5R1
|
cytochrome b5 reductase 1 |
| chr14_+_56584414 | 0.04 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr17_+_56833184 | 0.03 |
ENST00000308249.2
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr9_-_139361503 | 0.03 |
ENST00000453963.1
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr6_+_1080164 | 0.03 |
ENST00000314040.1
|
AL033381.1
|
Uncharacterized protein; cDNA FLJ34594 fis, clone KIDNE2009109 |
| chr5_+_156887027 | 0.03 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chrX_-_15333736 | 0.03 |
ENST00000380470.3
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chrX_+_36053908 | 0.03 |
ENST00000378660.2
|
CHDC2
|
calponin homology domain containing 2 |
| chr12_+_101869096 | 0.03 |
ENST00000551346.1
|
SPIC
|
Spi-C transcription factor (Spi-1/PU.1 related) |
| chr3_+_41241596 | 0.03 |
ENST00000450969.1
|
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr12_+_109826524 | 0.03 |
ENST00000431443.2
|
MYO1H
|
myosin IH |
| chr7_-_92855762 | 0.03 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr5_+_118812294 | 0.03 |
ENST00000509514.1
|
HSD17B4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr3_+_38206975 | 0.03 |
ENST00000446845.1
ENST00000311806.3 |
OXSR1
|
oxidative stress responsive 1 |
| chr7_+_150065278 | 0.03 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr16_+_30212378 | 0.03 |
ENST00000569485.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr20_+_56964169 | 0.03 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr10_+_89420706 | 0.03 |
ENST00000427144.2
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr2_-_24270217 | 0.03 |
ENST00000295148.4
ENST00000406895.3 |
C2orf44
|
chromosome 2 open reading frame 44 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.2 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.4 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.2 | GO:0044145 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.5 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0060723 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.0 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.0 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.0 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.2 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.4 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.0 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.0 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |