Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
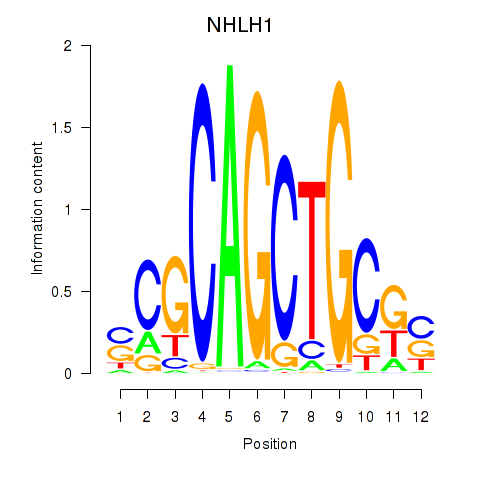
Results for NHLH1
Z-value: 0.89
Transcription factors associated with NHLH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NHLH1
|
ENSG00000171786.5 | nescient helix-loop-helix 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NHLH1 | hg19_v2_chr1_+_160336851_160336868 | 0.20 | 7.1e-01 | Click! |
Activity profile of NHLH1 motif
Sorted Z-values of NHLH1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_39258461 | 1.13 |
ENST00000440582.1
|
KRTAP4-16P
|
keratin associated protein 4-16, pseudogene |
| chr17_+_38171614 | 0.70 |
ENST00000583218.1
ENST00000394149.3 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr20_+_44637526 | 0.64 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr6_-_160148356 | 0.63 |
ENST00000401980.3
ENST00000545162.1 |
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr6_+_31895467 | 0.54 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr6_+_31895480 | 0.49 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr1_-_38273840 | 0.43 |
ENST00000373044.2
|
YRDC
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chrX_+_70521584 | 0.40 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr12_-_58145889 | 0.39 |
ENST00000547853.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr10_+_12391685 | 0.38 |
ENST00000378845.1
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr11_+_71238313 | 0.34 |
ENST00000398536.4
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr16_-_2770216 | 0.32 |
ENST00000302641.3
|
PRSS27
|
protease, serine 27 |
| chr4_-_2758015 | 0.32 |
ENST00000510267.1
ENST00000503235.1 ENST00000315423.7 |
TNIP2
|
TNFAIP3 interacting protein 2 |
| chr4_-_80994210 | 0.32 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr12_-_86650077 | 0.30 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr1_-_2461684 | 0.30 |
ENST00000378453.3
|
HES5
|
hes family bHLH transcription factor 5 |
| chr6_-_30712313 | 0.30 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr11_-_1619524 | 0.30 |
ENST00000412090.1
|
KRTAP5-2
|
keratin associated protein 5-2 |
| chr19_+_39989535 | 0.29 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr11_+_61276214 | 0.29 |
ENST00000378075.2
|
LRRC10B
|
leucine rich repeat containing 10B |
| chr12_-_113658892 | 0.29 |
ENST00000299732.2
ENST00000416617.2 |
IQCD
|
IQ motif containing D |
| chr16_+_23847339 | 0.29 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr4_+_89299994 | 0.29 |
ENST00000264346.7
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr6_-_38607628 | 0.28 |
ENST00000498633.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr6_+_126112001 | 0.28 |
ENST00000392477.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr1_-_119682812 | 0.28 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr2_+_58655461 | 0.28 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr2_-_230135937 | 0.28 |
ENST00000392054.3
ENST00000409462.1 ENST00000392055.3 |
PID1
|
phosphotyrosine interaction domain containing 1 |
| chr5_-_138739739 | 0.27 |
ENST00000514983.1
ENST00000507779.2 ENST00000451821.2 ENST00000450845.2 ENST00000509959.1 ENST00000302091.5 |
SPATA24
|
spermatogenesis associated 24 |
| chr4_+_89299885 | 0.27 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr7_-_131241361 | 0.27 |
ENST00000378555.3
ENST00000322985.9 ENST00000541194.1 ENST00000537928.1 |
PODXL
|
podocalyxin-like |
| chr7_-_6388389 | 0.26 |
ENST00000578372.1
|
FAM220A
|
family with sequence similarity 220, member A |
| chrX_-_19905577 | 0.26 |
ENST00000379697.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr9_-_112970436 | 0.25 |
ENST00000400613.4
|
C9orf152
|
chromosome 9 open reading frame 152 |
| chr9_-_117880477 | 0.25 |
ENST00000534839.1
ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC
|
tenascin C |
| chr12_-_54813229 | 0.24 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr2_-_96811170 | 0.24 |
ENST00000288943.4
|
DUSP2
|
dual specificity phosphatase 2 |
| chr6_+_126112074 | 0.23 |
ENST00000453302.1
ENST00000417494.1 ENST00000229634.9 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr6_-_75915757 | 0.23 |
ENST00000322507.8
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr22_-_24989014 | 0.23 |
ENST00000318753.8
|
FAM211B
|
family with sequence similarity 211, member B |
| chrX_-_43832711 | 0.23 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chr7_-_127671674 | 0.22 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr10_-_101380121 | 0.22 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr16_+_23847267 | 0.22 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr14_+_64971438 | 0.21 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr8_-_125740514 | 0.21 |
ENST00000325064.5
ENST00000518547.1 |
MTSS1
|
metastasis suppressor 1 |
| chr7_-_77045617 | 0.21 |
ENST00000257626.7
|
GSAP
|
gamma-secretase activating protein |
| chr16_-_88772670 | 0.21 |
ENST00000562544.1
|
RNF166
|
ring finger protein 166 |
| chr1_+_20915409 | 0.21 |
ENST00000375071.3
|
CDA
|
cytidine deaminase |
| chr1_+_13910479 | 0.21 |
ENST00000509009.1
|
PDPN
|
podoplanin |
| chr20_+_44036620 | 0.20 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr14_-_24732403 | 0.20 |
ENST00000206765.6
|
TGM1
|
transglutaminase 1 |
| chr1_+_226411319 | 0.20 |
ENST00000542034.1
ENST00000366810.5 |
MIXL1
|
Mix paired-like homeobox |
| chr3_+_167453026 | 0.20 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr2_+_196522032 | 0.20 |
ENST00000418005.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chrX_+_150151752 | 0.20 |
ENST00000325307.7
|
HMGB3
|
high mobility group box 3 |
| chr16_-_90085824 | 0.20 |
ENST00000002501.6
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1 |
| chr7_-_766879 | 0.20 |
ENST00000537384.1
ENST00000417852.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr15_+_91427726 | 0.19 |
ENST00000452243.1
|
FES
|
feline sarcoma oncogene |
| chr1_+_13910194 | 0.19 |
ENST00000376057.4
ENST00000510906.1 |
PDPN
|
podoplanin |
| chr12_+_90102729 | 0.19 |
ENST00000605386.1
|
LINC00936
|
long intergenic non-protein coding RNA 936 |
| chr22_+_32340481 | 0.19 |
ENST00000397492.1
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr22_+_32340447 | 0.19 |
ENST00000248975.5
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr5_+_52776228 | 0.18 |
ENST00000256759.3
|
FST
|
follistatin |
| chr15_-_102264619 | 0.18 |
ENST00000335968.3
|
TARSL2
|
threonyl-tRNA synthetase-like 2 |
| chr20_-_56100155 | 0.18 |
ENST00000423479.3
ENST00000502686.2 ENST00000433949.3 ENST00000539382.1 ENST00000608903.1 |
CTCFL
|
CCCTC-binding factor (zinc finger protein)-like |
| chr6_-_43543702 | 0.18 |
ENST00000265351.7
|
XPO5
|
exportin 5 |
| chr4_-_80994619 | 0.18 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chrX_-_19905703 | 0.17 |
ENST00000397821.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr1_+_32538520 | 0.17 |
ENST00000438825.1
ENST00000456834.2 ENST00000373634.4 ENST00000427288.1 |
TMEM39B
|
transmembrane protein 39B |
| chr5_-_178772424 | 0.17 |
ENST00000251582.7
ENST00000274609.5 |
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr11_+_110001723 | 0.17 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr1_-_8075693 | 0.17 |
ENST00000467067.1
|
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr15_-_60884706 | 0.17 |
ENST00000449337.2
|
RORA
|
RAR-related orphan receptor A |
| chr14_-_24732368 | 0.17 |
ENST00000544573.1
|
TGM1
|
transglutaminase 1 |
| chr3_+_49711777 | 0.17 |
ENST00000442186.1
ENST00000438011.1 ENST00000457042.1 |
APEH
|
acylaminoacyl-peptide hydrolase |
| chr1_+_32538492 | 0.17 |
ENST00000336294.5
|
TMEM39B
|
transmembrane protein 39B |
| chr4_+_87856191 | 0.17 |
ENST00000503477.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chrX_-_68385354 | 0.17 |
ENST00000361478.1
|
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr3_+_49711391 | 0.16 |
ENST00000296456.5
ENST00000449966.1 |
APEH
|
acylaminoacyl-peptide hydrolase |
| chr10_+_103348031 | 0.16 |
ENST00000370151.4
ENST00000370147.1 ENST00000370148.2 |
DPCD
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chrX_+_49126294 | 0.16 |
ENST00000466508.1
ENST00000438316.1 ENST00000055335.6 ENST00000495799.1 |
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F |
| chr1_-_22469459 | 0.16 |
ENST00000290167.6
|
WNT4
|
wingless-type MMTV integration site family, member 4 |
| chr5_-_16936340 | 0.16 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr14_-_64971288 | 0.16 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr1_+_14075903 | 0.16 |
ENST00000343137.4
ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr2_+_137523086 | 0.16 |
ENST00000409968.1
|
THSD7B
|
thrombospondin, type I, domain containing 7B |
| chr6_+_30848829 | 0.16 |
ENST00000508317.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr17_-_1012305 | 0.16 |
ENST00000291107.2
|
ABR
|
active BCR-related |
| chr17_-_54991369 | 0.16 |
ENST00000537230.1
|
TRIM25
|
tripartite motif containing 25 |
| chr12_+_27932803 | 0.15 |
ENST00000381271.2
|
KLHL42
|
kelch-like family member 42 |
| chr2_+_48541776 | 0.15 |
ENST00000413569.1
ENST00000340553.3 |
FOXN2
|
forkhead box N2 |
| chr11_+_62556596 | 0.15 |
ENST00000526546.1
|
TMEM179B
|
transmembrane protein 179B |
| chr6_+_30848740 | 0.15 |
ENST00000505534.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr13_-_28194541 | 0.15 |
ENST00000316334.3
|
LNX2
|
ligand of numb-protein X 2 |
| chr17_-_79792909 | 0.15 |
ENST00000330261.4
ENST00000570394.1 |
PPP1R27
|
protein phosphatase 1, regulatory subunit 27 |
| chr6_+_30848557 | 0.15 |
ENST00000460944.2
ENST00000324771.8 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr6_-_131211534 | 0.15 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr15_-_80215984 | 0.15 |
ENST00000485386.1
ENST00000479961.1 |
ST20
ST20-MTHFS
|
suppressor of tumorigenicity 20 ST20-MTHFS readthrough |
| chr19_-_47104118 | 0.15 |
ENST00000593888.1
ENST00000602017.1 |
AC011551.3
PPP5D1
|
Uncharacterized protein PPP5 tetratricopeptide repeat domain containing 1 |
| chr2_+_191513789 | 0.15 |
ENST00000409581.1
|
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chrX_-_68385274 | 0.15 |
ENST00000374584.3
ENST00000590146.1 |
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr20_+_35234137 | 0.14 |
ENST00000344795.3
ENST00000373852.5 |
C20orf24
|
chromosome 20 open reading frame 24 |
| chr2_+_65663812 | 0.14 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr20_+_44036900 | 0.14 |
ENST00000443296.1
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr2_-_70475586 | 0.14 |
ENST00000416149.2
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr1_-_38218577 | 0.14 |
ENST00000540011.1
|
EPHA10
|
EPH receptor A10 |
| chr6_-_43596899 | 0.14 |
ENST00000307126.5
ENST00000452781.1 |
GTPBP2
|
GTP binding protein 2 |
| chr11_-_71293921 | 0.14 |
ENST00000398530.1
|
KRTAP5-11
|
keratin associated protein 5-11 |
| chr3_+_99833755 | 0.14 |
ENST00000489081.1
|
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr15_+_91427642 | 0.14 |
ENST00000328850.3
ENST00000414248.2 |
FES
|
feline sarcoma oncogene |
| chr19_+_36024310 | 0.14 |
ENST00000222286.4
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr3_-_45187843 | 0.14 |
ENST00000296129.1
ENST00000425231.2 |
CDCP1
|
CUB domain containing protein 1 |
| chr7_-_767249 | 0.14 |
ENST00000403562.1
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr1_-_229694406 | 0.14 |
ENST00000344517.4
|
ABCB10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr19_+_16187085 | 0.14 |
ENST00000300933.4
|
TPM4
|
tropomyosin 4 |
| chr21_+_35445811 | 0.14 |
ENST00000399312.2
|
MRPS6
|
mitochondrial ribosomal protein S6 |
| chr12_+_113659234 | 0.14 |
ENST00000551096.1
ENST00000551099.1 ENST00000335509.6 ENST00000552897.1 ENST00000550785.1 ENST00000549279.1 |
TPCN1
|
two pore segment channel 1 |
| chr3_-_62860704 | 0.14 |
ENST00000490353.2
|
CADPS
|
Ca++-dependent secretion activator |
| chr17_-_15554940 | 0.13 |
ENST00000455584.2
|
RP11-385D13.1
|
Uncharacterized protein |
| chr10_+_1102303 | 0.13 |
ENST00000381329.1
|
WDR37
|
WD repeat domain 37 |
| chr2_+_10442993 | 0.13 |
ENST00000423674.1
ENST00000307845.3 |
HPCAL1
|
hippocalcin-like 1 |
| chr11_+_62554860 | 0.13 |
ENST00000533861.1
ENST00000333449.4 |
TMEM179B
|
transmembrane protein 179B |
| chr1_-_207096529 | 0.13 |
ENST00000525793.1
ENST00000529560.1 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr12_-_110318130 | 0.13 |
ENST00000540772.1
|
GLTP
|
glycolipid transfer protein |
| chr11_-_74022658 | 0.13 |
ENST00000427714.2
ENST00000331597.4 |
P4HA3
|
prolyl 4-hydroxylase, alpha polypeptide III |
| chr20_-_43438912 | 0.13 |
ENST00000541604.2
ENST00000372851.3 |
RIMS4
|
regulating synaptic membrane exocytosis 4 |
| chr10_+_112257596 | 0.13 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr14_+_73525265 | 0.13 |
ENST00000525161.1
|
RBM25
|
RNA binding motif protein 25 |
| chr14_+_31343747 | 0.13 |
ENST00000216361.4
ENST00000396618.3 ENST00000475087.1 |
COCH
|
cochlin |
| chr3_-_73673991 | 0.13 |
ENST00000308537.4
ENST00000263666.4 |
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr12_-_113658826 | 0.12 |
ENST00000546692.1
|
IQCD
|
IQ motif containing D |
| chr5_+_49962772 | 0.12 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr20_-_48770244 | 0.12 |
ENST00000371650.5
ENST00000371652.4 ENST00000557021.1 |
TMEM189
|
transmembrane protein 189 |
| chr6_+_30848771 | 0.12 |
ENST00000503180.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr10_-_103347883 | 0.12 |
ENST00000339310.3
ENST00000370158.3 ENST00000299206.4 ENST00000456836.2 ENST00000413344.1 ENST00000429502.1 ENST00000430045.1 ENST00000370172.1 ENST00000436284.2 ENST00000370162.3 |
POLL
|
polymerase (DNA directed), lambda |
| chr6_+_4776580 | 0.12 |
ENST00000397588.3
|
CDYL
|
chromodomain protein, Y-like |
| chr1_-_119683251 | 0.12 |
ENST00000369426.5
ENST00000235521.4 |
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr19_+_10217364 | 0.12 |
ENST00000430370.1
|
PPAN
|
peter pan homolog (Drosophila) |
| chr17_-_73266616 | 0.12 |
ENST00000579194.1
ENST00000581777.1 |
MIF4GD
|
MIF4G domain containing |
| chr3_+_5020801 | 0.12 |
ENST00000256495.3
|
BHLHE40
|
basic helix-loop-helix family, member e40 |
| chr12_+_57984965 | 0.12 |
ENST00000540759.2
ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr2_+_69001913 | 0.12 |
ENST00000409030.3
ENST00000409220.1 |
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr6_-_43337180 | 0.12 |
ENST00000318149.3
ENST00000361428.2 |
ZNF318
|
zinc finger protein 318 |
| chr5_+_52776449 | 0.12 |
ENST00000396947.3
|
FST
|
follistatin |
| chr1_-_19536744 | 0.12 |
ENST00000375267.2
ENST00000375217.2 ENST00000375226.2 ENST00000375254.3 |
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr15_+_91427691 | 0.12 |
ENST00000559355.1
ENST00000394302.1 |
FES
|
feline sarcoma oncogene |
| chr8_+_128747757 | 0.12 |
ENST00000517291.1
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr12_+_652294 | 0.11 |
ENST00000322843.3
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr8_+_86376081 | 0.11 |
ENST00000285379.5
|
CA2
|
carbonic anhydrase II |
| chr16_+_67562702 | 0.11 |
ENST00000379312.3
ENST00000042381.4 ENST00000540839.3 |
FAM65A
|
family with sequence similarity 65, member A |
| chr17_-_8301132 | 0.11 |
ENST00000399398.2
|
RNF222
|
ring finger protein 222 |
| chr6_-_44095183 | 0.11 |
ENST00000372014.3
|
MRPL14
|
mitochondrial ribosomal protein L14 |
| chr12_-_110318226 | 0.11 |
ENST00000544393.1
|
GLTP
|
glycolipid transfer protein |
| chr14_+_90863327 | 0.11 |
ENST00000356978.4
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr1_-_92351666 | 0.11 |
ENST00000465892.2
ENST00000417833.2 |
TGFBR3
|
transforming growth factor, beta receptor III |
| chr19_+_47104553 | 0.11 |
ENST00000598871.1
ENST00000594523.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr8_+_128748308 | 0.11 |
ENST00000377970.2
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr19_-_59070239 | 0.11 |
ENST00000595957.1
ENST00000253023.3 |
UBE2M
|
ubiquitin-conjugating enzyme E2M |
| chr11_+_17756279 | 0.11 |
ENST00000265969.6
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chr7_-_105925558 | 0.11 |
ENST00000222553.3
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr12_-_118406777 | 0.11 |
ENST00000339824.5
|
KSR2
|
kinase suppressor of ras 2 |
| chr8_+_24772455 | 0.11 |
ENST00000433454.2
|
NEFM
|
neurofilament, medium polypeptide |
| chr7_+_20370300 | 0.11 |
ENST00000537992.1
|
ITGB8
|
integrin, beta 8 |
| chr20_+_32399093 | 0.11 |
ENST00000217402.2
|
CHMP4B
|
charged multivesicular body protein 4B |
| chr4_+_75311019 | 0.11 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr6_-_3157760 | 0.11 |
ENST00000333628.3
|
TUBB2A
|
tubulin, beta 2A class IIa |
| chr6_+_31730773 | 0.11 |
ENST00000415669.2
ENST00000425424.1 |
SAPCD1
|
suppressor APC domain containing 1 |
| chr12_-_2944184 | 0.10 |
ENST00000337508.4
|
NRIP2
|
nuclear receptor interacting protein 2 |
| chrX_+_84498989 | 0.10 |
ENST00000395402.1
|
ZNF711
|
zinc finger protein 711 |
| chr16_+_69373471 | 0.10 |
ENST00000569637.2
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr16_+_330581 | 0.10 |
ENST00000219409.3
|
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr9_-_104357277 | 0.10 |
ENST00000374806.1
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta |
| chr9_+_2017420 | 0.10 |
ENST00000439732.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr16_+_69373323 | 0.10 |
ENST00000254940.5
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr17_-_27278304 | 0.10 |
ENST00000577226.1
|
PHF12
|
PHD finger protein 12 |
| chr11_-_75141206 | 0.10 |
ENST00000376292.4
|
KLHL35
|
kelch-like family member 35 |
| chr19_-_55953704 | 0.10 |
ENST00000416792.1
|
SHISA7
|
shisa family member 7 |
| chr20_+_35234223 | 0.10 |
ENST00000342422.3
|
C20orf24
|
chromosome 20 open reading frame 24 |
| chr1_+_15272271 | 0.10 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr14_+_70346125 | 0.10 |
ENST00000361956.3
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chr14_-_89021077 | 0.10 |
ENST00000556564.1
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr14_+_24584056 | 0.10 |
ENST00000561001.1
|
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr14_-_101053739 | 0.10 |
ENST00000554140.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr6_-_154831779 | 0.10 |
ENST00000607772.1
|
CNKSR3
|
CNKSR family member 3 |
| chr19_+_35521572 | 0.10 |
ENST00000262631.5
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr6_+_129204337 | 0.10 |
ENST00000421865.2
|
LAMA2
|
laminin, alpha 2 |
| chr16_-_67694129 | 0.10 |
ENST00000602320.1
|
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr20_-_48770174 | 0.09 |
ENST00000341698.2
|
TMEM189-UBE2V1
|
TMEM189-UBE2V1 readthrough |
| chr19_+_16186903 | 0.09 |
ENST00000588507.1
|
TPM4
|
tropomyosin 4 |
| chr1_+_157963063 | 0.09 |
ENST00000360089.4
ENST00000368173.3 ENST00000392272.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr18_-_5544241 | 0.09 |
ENST00000341928.2
ENST00000540638.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr2_+_103236004 | 0.09 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr1_+_32687971 | 0.09 |
ENST00000373586.1
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I |
| chr2_+_54683419 | 0.09 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr3_+_167453493 | 0.09 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr16_-_89883015 | 0.09 |
ENST00000563673.1
ENST00000389301.3 ENST00000568369.1 ENST00000534992.1 ENST00000389302.3 ENST00000543736.1 |
FANCA
|
Fanconi anemia, complementation group A |
| chr6_+_30851840 | 0.09 |
ENST00000511510.1
ENST00000376569.3 ENST00000376575.3 ENST00000376570.4 ENST00000446312.1 ENST00000504927.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr13_+_49794474 | 0.09 |
ENST00000218721.1
ENST00000398307.1 |
MLNR
|
motilin receptor |
| chr18_+_3449821 | 0.09 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NHLH1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 0.4 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.6 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.4 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.3 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.2 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.1 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.6 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.2 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.2 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.1 | 0.2 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.1 | 0.2 | GO:0060381 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.1 | 0.3 | GO:2001170 | positive regulation of fat cell proliferation(GO:0070346) negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 | 0.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.7 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.2 | GO:0061183 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) renal vesicle induction(GO:0072034) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.4 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.2 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.2 | GO:0090096 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.4 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.4 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.2 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0042938 | positive regulation of cellular pH reduction(GO:0032849) dipeptide transport(GO:0042938) |
| 0.0 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.1 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.1 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.3 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.3 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.2 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.0 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.1 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.0 | 0.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.0 | GO:0072186 | metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.2 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.0 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.4 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.2 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.3 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 | 0.1 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.3 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.2 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.1 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) |
| 0.0 | 0.0 | GO:0035696 | egg activation(GO:0007343) monocyte extravasation(GO:0035696) regulation of fertilization(GO:0080154) activation of meiosis(GO:0090427) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.2 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0032661 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) regulation of interleukin-18 production(GO:0032661) negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.2 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.1 | 0.2 | GO:0042565 | nuclear RNA export factor complex(GO:0042272) RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.6 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.2 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.1 | 0.2 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.2 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 1.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.0 | 0.1 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 0.1 | GO:0098519 | phosphoserine phosphatase activity(GO:0004647) nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |