Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
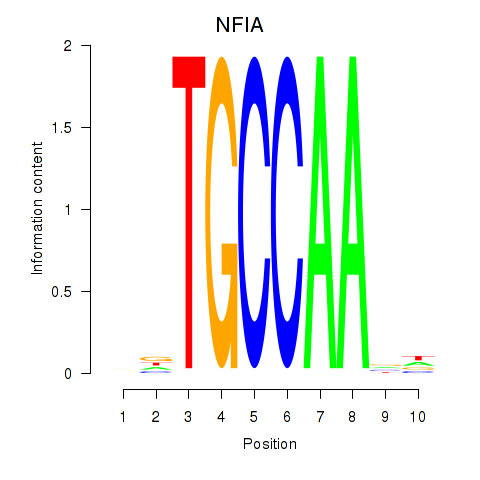
Results for NFIA
Z-value: 0.98
Transcription factors associated with NFIA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFIA
|
ENSG00000162599.11 | nuclear factor I A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFIA | hg19_v2_chr1_+_61548225_61548299 | -0.58 | 2.3e-01 | Click! |
Activity profile of NFIA motif
Sorted Z-values of NFIA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_17598506 | 1.19 |
ENST00000451792.1
|
AC017060.1
|
AC017060.1 |
| chr7_-_41742697 | 0.89 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr11_+_64004888 | 0.58 |
ENST00000541681.1
|
VEGFB
|
vascular endothelial growth factor B |
| chr12_-_15038779 | 0.53 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr2_-_214017151 | 0.52 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr15_-_79576156 | 0.47 |
ENST00000560452.1
ENST00000560872.1 ENST00000560732.1 ENST00000559979.1 ENST00000560533.1 ENST00000559225.1 |
RP11-17L5.4
|
RP11-17L5.4 |
| chr2_+_173955327 | 0.46 |
ENST00000422149.1
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr14_+_77582905 | 0.44 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr9_+_108424738 | 0.41 |
ENST00000334077.3
|
TAL2
|
T-cell acute lymphocytic leukemia 2 |
| chr12_-_53207842 | 0.39 |
ENST00000458244.2
|
KRT4
|
keratin 4 |
| chrX_-_133792480 | 0.36 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr8_+_40010989 | 0.35 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr2_-_31637560 | 0.34 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr17_+_65375082 | 0.34 |
ENST00000584471.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr1_+_41447609 | 0.32 |
ENST00000543104.1
|
CTPS1
|
CTP synthase 1 |
| chr7_-_21985489 | 0.30 |
ENST00000356195.5
ENST00000447180.1 ENST00000373934.4 ENST00000457951.1 |
CDCA7L
|
cell division cycle associated 7-like |
| chr21_-_28215332 | 0.29 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr17_+_1733251 | 0.29 |
ENST00000570451.1
|
RPA1
|
replication protein A1, 70kDa |
| chr17_-_47786375 | 0.28 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr5_-_137374288 | 0.27 |
ENST00000514310.1
|
FAM13B
|
family with sequence similarity 13, member B |
| chr10_-_106240032 | 0.26 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr4_-_87770416 | 0.26 |
ENST00000273905.6
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter), member 6 |
| chr15_+_36994210 | 0.25 |
ENST00000562489.1
|
C15orf41
|
chromosome 15 open reading frame 41 |
| chr7_-_151330218 | 0.25 |
ENST00000476632.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr6_+_43968306 | 0.24 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr17_-_15168624 | 0.24 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr5_-_55008136 | 0.24 |
ENST00000503891.1
ENST00000507109.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr3_+_109128961 | 0.23 |
ENST00000489670.1
|
RP11-702L6.4
|
RP11-702L6.4 |
| chr3_+_111718173 | 0.23 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr14_-_98444386 | 0.23 |
ENST00000556462.1
ENST00000556138.1 |
C14orf64
|
chromosome 14 open reading frame 64 |
| chr3_+_142342240 | 0.23 |
ENST00000497199.1
|
PLS1
|
plastin 1 |
| chr1_+_209878182 | 0.22 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr3_-_155011483 | 0.22 |
ENST00000489090.1
|
RP11-451G4.2
|
RP11-451G4.2 |
| chr1_-_204119009 | 0.22 |
ENST00000444817.1
|
ETNK2
|
ethanolamine kinase 2 |
| chr16_+_82090028 | 0.22 |
ENST00000568090.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr3_-_93747425 | 0.21 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr7_-_7782204 | 0.20 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr3_+_172361483 | 0.20 |
ENST00000598405.1
|
AC007919.2
|
HCG1787166; PRO1163; Uncharacterized protein |
| chr9_-_5437818 | 0.20 |
ENST00000223864.2
|
PLGRKT
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr11_-_122931881 | 0.20 |
ENST00000526110.1
ENST00000227378.3 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr6_-_113754604 | 0.20 |
ENST00000421737.1
|
RP1-124C6.1
|
RP1-124C6.1 |
| chr2_-_113542063 | 0.19 |
ENST00000263339.3
|
IL1A
|
interleukin 1, alpha |
| chr2_-_61244550 | 0.19 |
ENST00000421319.1
|
PUS10
|
pseudouridylate synthase 10 |
| chr5_+_53751445 | 0.19 |
ENST00000302005.1
|
HSPB3
|
heat shock 27kDa protein 3 |
| chr11_-_128392085 | 0.19 |
ENST00000526145.2
ENST00000531611.1 ENST00000319397.6 ENST00000345075.4 ENST00000535549.1 |
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr1_-_205912577 | 0.19 |
ENST00000367135.3
ENST00000367134.2 |
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr16_-_55866997 | 0.19 |
ENST00000360526.3
ENST00000361503.4 |
CES1
|
carboxylesterase 1 |
| chr2_+_152214098 | 0.18 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr2_+_54684327 | 0.18 |
ENST00000389980.3
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr8_+_86851932 | 0.18 |
ENST00000517368.1
|
CTA-392E5.1
|
CTA-392E5.1 |
| chr21_-_35340759 | 0.18 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chr17_+_7461580 | 0.18 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr4_+_153021899 | 0.18 |
ENST00000509332.1
ENST00000504144.1 ENST00000499452.2 |
RP11-18H21.1
|
RP11-18H21.1 |
| chr16_-_19725899 | 0.18 |
ENST00000567367.1
|
KNOP1
|
lysine-rich nucleolar protein 1 |
| chr11_+_126262027 | 0.18 |
ENST00000526311.1
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr8_+_32579321 | 0.18 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr8_+_32579271 | 0.17 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr4_-_48271802 | 0.17 |
ENST00000381501.3
|
TEC
|
tec protein tyrosine kinase |
| chr7_-_99381798 | 0.17 |
ENST00000415003.1
ENST00000354593.2 |
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr10_+_102821551 | 0.17 |
ENST00000370200.5
|
KAZALD1
|
Kazal-type serine peptidase inhibitor domain 1 |
| chr2_+_234545148 | 0.17 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr1_-_41950342 | 0.17 |
ENST00000372587.4
|
EDN2
|
endothelin 2 |
| chr12_+_82752275 | 0.17 |
ENST00000248306.3
|
METTL25
|
methyltransferase like 25 |
| chr1_+_112016414 | 0.17 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr1_+_95975672 | 0.17 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr3_-_52488048 | 0.17 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chr15_+_41549105 | 0.17 |
ENST00000560965.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr7_+_129015484 | 0.16 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr13_+_78109804 | 0.16 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr4_+_169633310 | 0.16 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr11_+_34654011 | 0.16 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr16_-_55867146 | 0.16 |
ENST00000422046.2
|
CES1
|
carboxylesterase 1 |
| chr1_-_243417762 | 0.16 |
ENST00000522191.1
|
CEP170
|
centrosomal protein 170kDa |
| chr22_+_25615489 | 0.16 |
ENST00000398215.2
|
CRYBB2
|
crystallin, beta B2 |
| chr16_-_74455290 | 0.16 |
ENST00000339953.5
|
CLEC18B
|
C-type lectin domain family 18, member B |
| chr16_+_67840986 | 0.16 |
ENST00000561639.1
ENST00000567852.1 ENST00000565148.1 ENST00000388833.3 ENST00000561654.1 ENST00000431934.2 |
TSNAXIP1
|
translin-associated factor X interacting protein 1 |
| chr2_+_33701684 | 0.16 |
ENST00000442390.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr4_-_157892167 | 0.16 |
ENST00000541126.1
|
PDGFC
|
platelet derived growth factor C |
| chr20_+_43343476 | 0.16 |
ENST00000372868.2
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr3_-_123168551 | 0.16 |
ENST00000462833.1
|
ADCY5
|
adenylate cyclase 5 |
| chr13_-_46425865 | 0.16 |
ENST00000400405.2
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr2_-_200320768 | 0.15 |
ENST00000428695.1
|
SATB2
|
SATB homeobox 2 |
| chr5_-_43313574 | 0.15 |
ENST00000325110.6
ENST00000433297.2 |
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr19_+_13988061 | 0.15 |
ENST00000339133.5
ENST00000397555.2 |
NANOS3
|
nanos homolog 3 (Drosophila) |
| chrX_-_71497077 | 0.15 |
ENST00000373626.3
|
RPS4X
|
ribosomal protein S4, X-linked |
| chr4_+_146402346 | 0.15 |
ENST00000514778.1
ENST00000507594.1 |
SMAD1
|
SMAD family member 1 |
| chr16_+_70207686 | 0.15 |
ENST00000541793.2
ENST00000314151.8 ENST00000565806.1 ENST00000569347.2 ENST00000536907.2 |
CLEC18C
|
C-type lectin domain family 18, member C |
| chr18_+_55712915 | 0.15 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr2_-_231989808 | 0.15 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr9_+_5890802 | 0.15 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chrX_-_71497148 | 0.15 |
ENST00000316084.6
|
RPS4X
|
ribosomal protein S4, X-linked |
| chr9_+_84304628 | 0.15 |
ENST00000437181.1
|
RP11-154D17.1
|
RP11-154D17.1 |
| chr2_-_136288740 | 0.14 |
ENST00000264159.6
ENST00000536680.1 |
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr20_+_43343886 | 0.14 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr3_+_4535155 | 0.14 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr3_+_190105909 | 0.14 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr16_+_15737124 | 0.14 |
ENST00000396355.1
ENST00000396353.2 |
NDE1
|
nudE neurodevelopment protein 1 |
| chr11_+_15136462 | 0.14 |
ENST00000379556.3
ENST00000424273.1 |
INSC
|
inscuteable homolog (Drosophila) |
| chr17_+_9745786 | 0.14 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr16_-_89119355 | 0.14 |
ENST00000537498.1
|
CTD-2555A7.2
|
CTD-2555A7.2 |
| chr6_-_2971494 | 0.13 |
ENST00000380539.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr5_-_64777733 | 0.13 |
ENST00000381055.3
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr2_-_74645669 | 0.13 |
ENST00000518401.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr5_-_168006324 | 0.13 |
ENST00000522176.1
|
PANK3
|
pantothenate kinase 3 |
| chr17_-_39459103 | 0.13 |
ENST00000391353.1
|
KRTAP29-1
|
keratin associated protein 29-1 |
| chr2_+_179184955 | 0.13 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr8_+_55471630 | 0.13 |
ENST00000522001.1
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr1_-_154164534 | 0.13 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr2_+_159651821 | 0.13 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr9_+_130911770 | 0.13 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr19_+_42254885 | 0.13 |
ENST00000595740.1
|
CEACAM6
|
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
| chr14_-_69350920 | 0.13 |
ENST00000553290.1
|
ACTN1
|
actinin, alpha 1 |
| chr11_+_111749650 | 0.13 |
ENST00000528125.1
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr21_+_19617140 | 0.13 |
ENST00000299295.2
ENST00000338326.3 |
CHODL
|
chondrolectin |
| chr3_+_150264458 | 0.13 |
ENST00000487799.1
ENST00000460851.1 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr8_+_66619277 | 0.12 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr16_-_65106110 | 0.12 |
ENST00000562882.1
ENST00000567934.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr3_+_152017924 | 0.12 |
ENST00000465907.2
ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr10_-_126138622 | 0.12 |
ENST00000451024.3
|
NKX1-2
|
NK1 homeobox 2 |
| chr14_+_70234642 | 0.12 |
ENST00000555349.1
ENST00000554021.1 |
SRSF5
|
serine/arginine-rich splicing factor 5 |
| chr5_+_132208014 | 0.12 |
ENST00000296877.2
|
LEAP2
|
liver expressed antimicrobial peptide 2 |
| chr17_+_7462031 | 0.12 |
ENST00000380535.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr4_-_101439242 | 0.12 |
ENST00000296420.4
|
EMCN
|
endomucin |
| chr21_+_44394742 | 0.12 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr3_-_33260283 | 0.12 |
ENST00000412539.1
|
SUSD5
|
sushi domain containing 5 |
| chr2_-_111291587 | 0.12 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr2_-_227050079 | 0.12 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr4_+_71108300 | 0.12 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr11_+_3875757 | 0.12 |
ENST00000525403.1
|
STIM1
|
stromal interaction molecule 1 |
| chr2_+_135596180 | 0.12 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr3_+_72937182 | 0.12 |
ENST00000389617.4
|
GXYLT2
|
glucoside xylosyltransferase 2 |
| chr5_-_111093759 | 0.12 |
ENST00000509979.1
ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP
|
neuronal regeneration related protein |
| chr9_-_127710292 | 0.12 |
ENST00000421514.1
|
GOLGA1
|
golgin A1 |
| chr6_+_42989344 | 0.12 |
ENST00000244496.5
|
RRP36
|
ribosomal RNA processing 36 homolog (S. cerevisiae) |
| chr17_+_19314505 | 0.12 |
ENST00000461366.1
|
RNF112
|
ring finger protein 112 |
| chr3_+_150264555 | 0.12 |
ENST00000406576.3
ENST00000482093.1 ENST00000273435.5 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr14_-_98444438 | 0.12 |
ENST00000512901.2
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr8_+_101349823 | 0.11 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr3_+_10312604 | 0.11 |
ENST00000426850.1
|
TATDN2
|
TatD DNase domain containing 2 |
| chr11_-_45928830 | 0.11 |
ENST00000449465.1
|
C11orf94
|
chromosome 11 open reading frame 94 |
| chr3_-_131736593 | 0.11 |
ENST00000514999.1
|
CPNE4
|
copine IV |
| chr1_+_107683436 | 0.11 |
ENST00000370068.1
|
NTNG1
|
netrin G1 |
| chr8_-_121824374 | 0.11 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr6_-_133079022 | 0.11 |
ENST00000525289.1
ENST00000326499.6 |
VNN2
|
vanin 2 |
| chr12_+_57828521 | 0.11 |
ENST00000309668.2
|
INHBC
|
inhibin, beta C |
| chr5_+_36608280 | 0.11 |
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr8_+_9009296 | 0.11 |
ENST00000521718.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr12_+_56325231 | 0.11 |
ENST00000549368.1
|
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr19_-_13710678 | 0.11 |
ENST00000592864.1
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr1_-_144364246 | 0.11 |
ENST00000540273.1
|
PPIAL4B
|
peptidylprolyl isomerase A (cyclophilin A)-like 4B |
| chr1_+_149553003 | 0.11 |
ENST00000369222.3
|
PPIAL4C
|
peptidylprolyl isomerase A (cyclophilin A)-like 4C |
| chrX_-_106146547 | 0.11 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr5_-_43313269 | 0.10 |
ENST00000511774.1
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr11_-_61101247 | 0.10 |
ENST00000543627.1
|
DDB1
|
damage-specific DNA binding protein 1, 127kDa |
| chr14_-_75083313 | 0.10 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr6_+_30951487 | 0.10 |
ENST00000486149.2
ENST00000376296.3 |
MUC21
|
mucin 21, cell surface associated |
| chr17_+_1733276 | 0.10 |
ENST00000254719.5
|
RPA1
|
replication protein A1, 70kDa |
| chr1_+_68150744 | 0.10 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr4_-_114900831 | 0.10 |
ENST00000315366.7
|
ARSJ
|
arylsulfatase family, member J |
| chr11_-_65625678 | 0.10 |
ENST00000308162.5
|
CFL1
|
cofilin 1 (non-muscle) |
| chr2_+_102953608 | 0.10 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr11_+_28724129 | 0.10 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr3_+_159570722 | 0.10 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr3_+_32737027 | 0.10 |
ENST00000454516.2
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr2_-_242211359 | 0.10 |
ENST00000444092.1
|
HDLBP
|
high density lipoprotein binding protein |
| chr15_+_48051920 | 0.10 |
ENST00000559196.1
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr13_+_102104952 | 0.10 |
ENST00000376180.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr7_-_154863264 | 0.10 |
ENST00000395731.2
ENST00000543018.1 |
HTR5A-AS1
|
HTR5A antisense RNA 1 |
| chr8_+_32579341 | 0.10 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr11_-_85397167 | 0.10 |
ENST00000316398.3
|
CCDC89
|
coiled-coil domain containing 89 |
| chr12_+_6977323 | 0.10 |
ENST00000462761.1
|
TPI1
|
triosephosphate isomerase 1 |
| chrX_-_19817869 | 0.10 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr11_-_111649015 | 0.10 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr20_+_43029911 | 0.10 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr9_+_117085336 | 0.10 |
ENST00000259396.8
ENST00000538816.1 |
ORM1
|
orosomucoid 1 |
| chr11_-_13461790 | 0.10 |
ENST00000530907.1
|
BTBD10
|
BTB (POZ) domain containing 10 |
| chr12_-_95510743 | 0.10 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr1_+_214163033 | 0.10 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chr10_-_101190202 | 0.10 |
ENST00000543866.1
ENST00000370508.5 |
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr1_+_110546700 | 0.10 |
ENST00000359172.3
ENST00000393614.4 |
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr5_+_36606992 | 0.10 |
ENST00000505202.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr11_-_118023594 | 0.10 |
ENST00000529878.1
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chrX_-_138287168 | 0.10 |
ENST00000436198.1
|
FGF13
|
fibroblast growth factor 13 |
| chr8_+_124194875 | 0.10 |
ENST00000522648.1
ENST00000276699.6 |
FAM83A
|
family with sequence similarity 83, member A |
| chr5_-_115890554 | 0.10 |
ENST00000509665.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr7_+_35756066 | 0.10 |
ENST00000449644.1
|
AC018647.3
|
AC018647.3 |
| chr7_+_114055052 | 0.09 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr15_+_67458357 | 0.09 |
ENST00000537194.2
|
SMAD3
|
SMAD family member 3 |
| chr1_-_160253998 | 0.09 |
ENST00000392220.2
|
PEX19
|
peroxisomal biogenesis factor 19 |
| chr1_-_237167718 | 0.09 |
ENST00000464121.2
|
MT1HL1
|
metallothionein 1H-like 1 |
| chrX_-_100604184 | 0.09 |
ENST00000372902.3
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr8_-_116504448 | 0.09 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr22_-_42086477 | 0.09 |
ENST00000402458.1
|
NHP2L1
|
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chr1_-_21978312 | 0.09 |
ENST00000359708.4
ENST00000290101.4 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr2_-_65659762 | 0.09 |
ENST00000440972.1
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr11_+_62556596 | 0.09 |
ENST00000526546.1
|
TMEM179B
|
transmembrane protein 179B |
| chr3_-_25824872 | 0.09 |
ENST00000308710.5
|
NGLY1
|
N-glycanase 1 |
| chr3_+_109128837 | 0.09 |
ENST00000497996.1
|
RP11-702L6.4
|
RP11-702L6.4 |
| chr11_+_111385497 | 0.09 |
ENST00000375618.4
ENST00000529167.1 ENST00000332814.6 |
C11orf88
|
chromosome 11 open reading frame 88 |
| chr21_+_27543175 | 0.09 |
ENST00000608591.1
ENST00000609365.1 |
AP000230.1
|
AP000230.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFIA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.3 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 0.3 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.3 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.1 | 0.6 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.3 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.1 | 0.5 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.2 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.2 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.2 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.2 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.0 | 0.2 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.2 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.4 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.3 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.2 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.0 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.2 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 0.0 | 0.4 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.0 | 0.1 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.2 | GO:2000782 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.2 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.2 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.5 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.0 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.0 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.3 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.0 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.0 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.2 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.3 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.0 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.0 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.0 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.0 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.2 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:0061589 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.3 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.0 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.0 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.6 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.2 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.1 | 0.3 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.2 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 0.2 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0031775 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.0 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |