Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
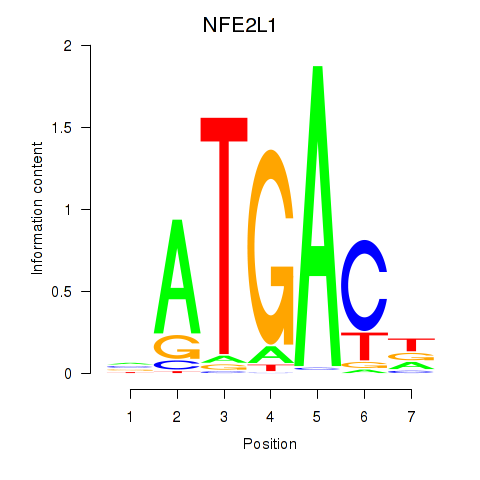
Results for NFE2L1
Z-value: 1.30
Transcription factors associated with NFE2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFE2L1
|
ENSG00000082641.11 | nuclear factor, erythroid 2 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFE2L1 | hg19_v2_chr17_+_46131912_46131952 | -0.92 | 8.6e-03 | Click! |
Activity profile of NFE2L1 motif
Sorted Z-values of NFE2L1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_74606223 | 0.87 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr2_+_113735575 | 0.86 |
ENST00000376489.2
ENST00000259205.4 |
IL36G
|
interleukin 36, gamma |
| chr2_-_25451065 | 0.80 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr6_+_13272904 | 0.67 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr1_+_81106951 | 0.67 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr16_+_31044413 | 0.61 |
ENST00000394998.1
|
STX4
|
syntaxin 4 |
| chr19_-_42192189 | 0.60 |
ENST00000401731.1
ENST00000338196.4 ENST00000006724.3 |
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr15_-_34502197 | 0.54 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr17_-_26220366 | 0.51 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr3_-_172019686 | 0.50 |
ENST00000596321.1
|
AC092964.2
|
Uncharacterized protein |
| chr6_-_133055815 | 0.48 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr18_-_2982869 | 0.46 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr8_+_55466915 | 0.46 |
ENST00000522711.2
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr6_-_152623231 | 0.45 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr1_-_173886491 | 0.43 |
ENST00000367698.3
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr3_-_186262166 | 0.42 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr2_+_149974684 | 0.42 |
ENST00000450639.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr17_+_57274914 | 0.41 |
ENST00000582004.1
ENST00000577660.1 |
PRR11
CTD-2510F5.6
|
proline rich 11 Uncharacterized protein |
| chr9_-_34665983 | 0.41 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr8_-_95220775 | 0.40 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr3_-_114790179 | 0.40 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr8_+_55467072 | 0.39 |
ENST00000602362.1
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr9_-_140082983 | 0.39 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr7_+_141463897 | 0.39 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr6_-_33037019 | 0.38 |
ENST00000437811.1
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr17_+_38119216 | 0.38 |
ENST00000301659.4
|
GSDMA
|
gasdermin A |
| chr15_+_101389945 | 0.37 |
ENST00000561231.1
ENST00000559331.1 ENST00000558254.1 |
RP11-66B24.2
|
RP11-66B24.2 |
| chr5_+_36608280 | 0.37 |
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr17_-_34207295 | 0.37 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr9_-_117853297 | 0.37 |
ENST00000542877.1
ENST00000537320.1 ENST00000341037.4 |
TNC
|
tenascin C |
| chr5_+_142286887 | 0.37 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr2_+_234160340 | 0.37 |
ENST00000417017.1
ENST00000392020.4 ENST00000392018.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr2_-_113594279 | 0.37 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr1_-_153113927 | 0.36 |
ENST00000368752.4
|
SPRR2B
|
small proline-rich protein 2B |
| chr1_-_12679171 | 0.36 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr16_-_67881588 | 0.36 |
ENST00000561593.1
ENST00000565114.1 |
CENPT
|
centromere protein T |
| chr6_+_31916733 | 0.36 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr11_+_64052692 | 0.36 |
ENST00000377702.4
|
GPR137
|
G protein-coupled receptor 137 |
| chr20_-_44516256 | 0.35 |
ENST00000372519.3
|
SPATA25
|
spermatogenesis associated 25 |
| chr1_-_219615984 | 0.35 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr5_-_176936844 | 0.34 |
ENST00000510380.1
ENST00000510898.1 ENST00000357198.4 |
DOK3
|
docking protein 3 |
| chr5_+_127039075 | 0.34 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chrX_-_84363974 | 0.34 |
ENST00000395409.3
ENST00000332921.5 ENST00000509231.1 |
SATL1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr11_-_10920838 | 0.34 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr22_+_30805086 | 0.33 |
ENST00000439838.1
ENST00000439023.3 |
RP4-539M6.19
|
Uncharacterized protein |
| chr3_-_100566492 | 0.33 |
ENST00000528490.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr17_-_57784755 | 0.32 |
ENST00000537860.1
ENST00000393038.2 ENST00000409433.2 |
PTRH2
|
peptidyl-tRNA hydrolase 2 |
| chr6_-_154751629 | 0.32 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr7_+_144015218 | 0.32 |
ENST00000408951.1
|
OR2A1
|
olfactory receptor, family 2, subfamily A, member 1 |
| chr5_-_162887054 | 0.32 |
ENST00000517501.1
|
NUDCD2
|
NudC domain containing 2 |
| chr6_-_31940065 | 0.32 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr6_+_143381979 | 0.32 |
ENST00000367598.5
ENST00000447498.1 ENST00000357847.4 ENST00000344492.5 ENST00000367596.1 ENST00000494282.2 ENST00000275235.4 |
AIG1
|
androgen-induced 1 |
| chr12_-_7596735 | 0.32 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr17_+_76210367 | 0.31 |
ENST00000592734.1
ENST00000587746.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr16_+_8814563 | 0.31 |
ENST00000425191.2
ENST00000569156.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr20_-_52612468 | 0.30 |
ENST00000422805.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr3_-_110612323 | 0.30 |
ENST00000383686.2
|
RP11-553A10.1
|
Uncharacterized protein |
| chr1_+_16330723 | 0.29 |
ENST00000329454.2
|
C1orf64
|
chromosome 1 open reading frame 64 |
| chr1_-_151319654 | 0.28 |
ENST00000430227.1
ENST00000412774.1 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr19_+_36545833 | 0.28 |
ENST00000401500.2
ENST00000270301.7 ENST00000427823.2 |
WDR62
|
WD repeat domain 62 |
| chr7_+_116593953 | 0.28 |
ENST00000397750.3
|
ST7-OT4
|
ST7 overlapping transcript 4 |
| chr7_+_872107 | 0.28 |
ENST00000405266.1
ENST00000401592.1 ENST00000403868.1 ENST00000425407.2 |
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr8_-_38386175 | 0.28 |
ENST00000437935.2
ENST00000358138.1 |
C8orf86
|
chromosome 8 open reading frame 86 |
| chr6_-_170151603 | 0.27 |
ENST00000366774.3
|
TCTE3
|
t-complex-associated-testis-expressed 3 |
| chr17_-_8021710 | 0.27 |
ENST00000380149.1
ENST00000448843.2 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chrM_+_7586 | 0.27 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr7_-_143929936 | 0.27 |
ENST00000391496.1
|
OR2A42
|
olfactory receptor, family 2, subfamily A, member 42 |
| chr19_+_36545781 | 0.27 |
ENST00000388999.3
|
WDR62
|
WD repeat domain 62 |
| chr2_+_234160217 | 0.27 |
ENST00000392017.4
ENST00000347464.5 ENST00000444735.1 ENST00000373525.5 ENST00000419681.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr10_-_51958906 | 0.27 |
ENST00000489640.1
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr11_+_64052266 | 0.26 |
ENST00000539851.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr20_+_57594309 | 0.26 |
ENST00000217133.1
|
TUBB1
|
tubulin, beta 1 class VI |
| chr1_-_11865982 | 0.26 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr17_-_40170506 | 0.26 |
ENST00000589773.1
ENST00000590348.1 |
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr7_-_41742697 | 0.26 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr4_+_71108300 | 0.26 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr6_-_43027105 | 0.26 |
ENST00000230413.5
ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2
|
mitochondrial ribosomal protein L2 |
| chr6_+_33043703 | 0.26 |
ENST00000418931.2
ENST00000535465.1 |
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr4_+_185734773 | 0.25 |
ENST00000508020.1
|
RP11-701P16.2
|
Uncharacterized protein |
| chr6_+_30295036 | 0.25 |
ENST00000376659.5
ENST00000428555.1 |
TRIM39
|
tripartite motif containing 39 |
| chr3_-_52488048 | 0.25 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chr19_-_39881669 | 0.24 |
ENST00000221266.7
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr9_+_125027127 | 0.24 |
ENST00000441707.1
ENST00000373723.5 ENST00000373729.1 |
MRRF
|
mitochondrial ribosome recycling factor |
| chr1_-_151319318 | 0.24 |
ENST00000436271.1
ENST00000450506.1 ENST00000422595.1 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr4_+_71384257 | 0.24 |
ENST00000339336.4
|
AMTN
|
amelotin |
| chr3_-_27764190 | 0.23 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr6_+_30029008 | 0.23 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr11_-_107729287 | 0.23 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr3_+_16926441 | 0.23 |
ENST00000418129.2
ENST00000396755.2 |
PLCL2
|
phospholipase C-like 2 |
| chr5_-_140700322 | 0.23 |
ENST00000313368.5
|
TAF7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr5_+_40909354 | 0.22 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr10_+_35464513 | 0.22 |
ENST00000494479.1
ENST00000463314.1 ENST00000342105.3 ENST00000495301.1 ENST00000463960.1 |
CREM
|
cAMP responsive element modulator |
| chr10_+_69865866 | 0.22 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr16_+_19467772 | 0.22 |
ENST00000219821.5
ENST00000561503.1 ENST00000564959.1 |
TMC5
|
transmembrane channel-like 5 |
| chr4_+_26324474 | 0.22 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chrX_+_119029800 | 0.22 |
ENST00000371431.3
ENST00000371423.2 ENST00000371425.4 ENST00000394594.2 |
AKAP14
|
A kinase (PRKA) anchor protein 14 |
| chr19_-_39881777 | 0.21 |
ENST00000595564.1
ENST00000221265.3 |
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr17_+_73663470 | 0.21 |
ENST00000583536.1
|
SAP30BP
|
SAP30 binding protein |
| chr22_+_32439019 | 0.21 |
ENST00000266088.4
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chrX_-_128782722 | 0.21 |
ENST00000427399.1
|
APLN
|
apelin |
| chr5_+_147582387 | 0.21 |
ENST00000325630.2
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr9_-_100684769 | 0.21 |
ENST00000455506.1
ENST00000375117.4 |
C9orf156
|
chromosome 9 open reading frame 156 |
| chr15_-_43559055 | 0.21 |
ENST00000220420.5
ENST00000349114.4 |
TGM5
|
transglutaminase 5 |
| chr19_-_36545649 | 0.21 |
ENST00000292894.1
|
THAP8
|
THAP domain containing 8 |
| chrX_+_47078069 | 0.21 |
ENST00000357227.4
ENST00000519758.1 ENST00000520893.1 ENST00000517426.1 |
CDK16
|
cyclin-dependent kinase 16 |
| chr12_+_57998400 | 0.21 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr19_+_44100632 | 0.21 |
ENST00000533118.1
|
ZNF576
|
zinc finger protein 576 |
| chr8_+_22853345 | 0.21 |
ENST00000522948.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr17_-_73663168 | 0.20 |
ENST00000578201.1
ENST00000423245.2 |
RECQL5
|
RecQ protein-like 5 |
| chr5_-_160279207 | 0.20 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr14_-_75536182 | 0.20 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr6_-_31939734 | 0.20 |
ENST00000375356.3
|
DXO
|
decapping exoribonuclease |
| chr1_+_43735646 | 0.19 |
ENST00000439858.1
|
TMEM125
|
transmembrane protein 125 |
| chr5_-_162887071 | 0.19 |
ENST00000302764.4
|
NUDCD2
|
NudC domain containing 2 |
| chr6_-_3157760 | 0.19 |
ENST00000333628.3
|
TUBB2A
|
tubulin, beta 2A class IIa |
| chr1_-_155211017 | 0.19 |
ENST00000536770.1
ENST00000368373.3 |
GBA
|
glucosidase, beta, acid |
| chr21_+_36041688 | 0.19 |
ENST00000360731.3
ENST00000349499.2 |
CLIC6
|
chloride intracellular channel 6 |
| chr19_+_47616682 | 0.19 |
ENST00000594526.1
|
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr11_+_64052454 | 0.19 |
ENST00000539833.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr15_+_94899183 | 0.19 |
ENST00000557742.1
|
MCTP2
|
multiple C2 domains, transmembrane 2 |
| chr22_-_37505449 | 0.19 |
ENST00000406725.1
|
TMPRSS6
|
transmembrane protease, serine 6 |
| chr1_+_45265897 | 0.19 |
ENST00000372201.4
|
PLK3
|
polo-like kinase 3 |
| chrX_+_70503037 | 0.19 |
ENST00000535149.1
|
NONO
|
non-POU domain containing, octamer-binding |
| chrX_+_100224676 | 0.19 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr4_-_147442982 | 0.19 |
ENST00000511374.1
ENST00000264986.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr17_+_7461580 | 0.18 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr2_-_87018784 | 0.18 |
ENST00000283635.3
ENST00000538832.1 |
CD8A
|
CD8a molecule |
| chrX_+_49126294 | 0.18 |
ENST00000466508.1
ENST00000438316.1 ENST00000055335.6 ENST00000495799.1 |
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F |
| chr1_+_198189921 | 0.18 |
ENST00000391974.3
|
NEK7
|
NIMA-related kinase 7 |
| chr6_+_144185573 | 0.18 |
ENST00000237275.6
ENST00000539295.1 |
ZC2HC1B
|
zinc finger, C2HC-type containing 1B |
| chr14_+_35761540 | 0.18 |
ENST00000261479.4
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr19_-_58485895 | 0.18 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr11_+_57531292 | 0.18 |
ENST00000524579.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr3_+_45730829 | 0.18 |
ENST00000438671.1
|
SACM1L
|
SAC1 suppressor of actin mutations 1-like (yeast) |
| chr15_-_45406385 | 0.18 |
ENST00000389039.6
|
DUOX2
|
dual oxidase 2 |
| chr3_+_63638372 | 0.18 |
ENST00000496807.1
|
SNTN
|
sentan, cilia apical structure protein |
| chr9_-_100684845 | 0.18 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156 |
| chr2_-_68547061 | 0.18 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr11_-_125366089 | 0.18 |
ENST00000366139.3
ENST00000278919.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr16_-_46797149 | 0.18 |
ENST00000536476.1
|
MYLK3
|
myosin light chain kinase 3 |
| chr9_+_140083099 | 0.18 |
ENST00000322310.5
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr1_+_67632083 | 0.17 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr8_-_86141245 | 0.17 |
ENST00000546501.1
|
RP11-219B4.5
|
Uncharacterized protein |
| chr3_-_98241358 | 0.17 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr3_+_45730912 | 0.17 |
ENST00000541314.1
|
SACM1L
|
SAC1 suppressor of actin mutations 1-like (yeast) |
| chr5_+_147582348 | 0.17 |
ENST00000514389.1
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr1_+_207039154 | 0.17 |
ENST00000367096.3
ENST00000391930.2 |
IL20
|
interleukin 20 |
| chr12_-_118628315 | 0.17 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr18_-_70535177 | 0.17 |
ENST00000327305.6
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr12_-_18890940 | 0.17 |
ENST00000543242.1
ENST00000539072.1 ENST00000541966.1 ENST00000266505.7 ENST00000447925.2 ENST00000435379.1 |
PLCZ1
|
phospholipase C, zeta 1 |
| chr17_-_39258461 | 0.17 |
ENST00000440582.1
|
KRTAP4-16P
|
keratin associated protein 4-16, pseudogene |
| chr11_+_7618413 | 0.17 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr7_+_6713376 | 0.17 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr4_-_40632605 | 0.17 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr1_-_119682812 | 0.16 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr4_-_103749179 | 0.16 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_+_131958436 | 0.16 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr19_+_4402659 | 0.16 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr19_+_18699599 | 0.16 |
ENST00000450195.2
|
C19orf60
|
chromosome 19 open reading frame 60 |
| chr14_-_21490653 | 0.16 |
ENST00000449431.2
|
NDRG2
|
NDRG family member 2 |
| chr1_+_149239529 | 0.16 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr11_-_108093329 | 0.16 |
ENST00000278612.8
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus |
| chr12_-_54813229 | 0.16 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr19_-_42721819 | 0.16 |
ENST00000336034.4
ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2
|
death effector domain containing 2 |
| chr15_+_71228826 | 0.16 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr10_-_49860525 | 0.16 |
ENST00000435790.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr4_+_71384300 | 0.16 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr8_+_24151553 | 0.16 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr15_+_39311432 | 0.16 |
ENST00000560709.1
|
RP11-62C7.2
|
RP11-62C7.2 |
| chr3_-_98241713 | 0.16 |
ENST00000502288.1
ENST00000512147.1 ENST00000510541.1 ENST00000503621.1 ENST00000511081.1 |
CLDND1
|
claudin domain containing 1 |
| chr2_-_46769694 | 0.16 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr1_+_222817629 | 0.16 |
ENST00000340535.7
|
MIA3
|
melanoma inhibitory activity family, member 3 |
| chr11_-_10920714 | 0.15 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr14_+_35761580 | 0.15 |
ENST00000553809.1
ENST00000555764.1 ENST00000556506.1 |
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr1_+_26517052 | 0.15 |
ENST00000338855.2
ENST00000456354.2 |
CATSPER4
|
cation channel, sperm associated 4 |
| chr7_-_11871815 | 0.15 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr15_-_45406348 | 0.15 |
ENST00000603300.1
|
DUOX2
|
dual oxidase 2 |
| chr11_-_126138808 | 0.15 |
ENST00000332118.6
ENST00000532259.1 |
SRPR
|
signal recognition particle receptor (docking protein) |
| chr4_+_165675197 | 0.15 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr6_+_143381594 | 0.15 |
ENST00000367601.4
|
AIG1
|
androgen-induced 1 |
| chr12_-_53228079 | 0.15 |
ENST00000330553.5
|
KRT79
|
keratin 79 |
| chr9_+_6757634 | 0.15 |
ENST00000543771.1
ENST00000401787.3 ENST00000381306.3 ENST00000381309.3 |
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr1_-_156265438 | 0.15 |
ENST00000362007.1
|
C1orf85
|
chromosome 1 open reading frame 85 |
| chr15_+_28624878 | 0.15 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr15_-_42840961 | 0.14 |
ENST00000563454.1
ENST00000397130.3 ENST00000570160.1 ENST00000323443.2 |
LRRC57
|
leucine rich repeat containing 57 |
| chr3_+_187957646 | 0.14 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr2_-_228244013 | 0.14 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr19_-_46142637 | 0.14 |
ENST00000590043.1
ENST00000589876.1 |
EML2
|
echinoderm microtubule associated protein like 2 |
| chr2_-_225266743 | 0.14 |
ENST00000409685.3
|
FAM124B
|
family with sequence similarity 124B |
| chr6_+_86195088 | 0.14 |
ENST00000437581.1
|
NT5E
|
5'-nucleotidase, ecto (CD73) |
| chr1_+_43613612 | 0.14 |
ENST00000335282.4
|
FAM183A
|
family with sequence similarity 183, member A |
| chr6_-_37467628 | 0.14 |
ENST00000373408.3
|
CCDC167
|
coiled-coil domain containing 167 |
| chr1_-_40562908 | 0.14 |
ENST00000527311.2
ENST00000449045.2 ENST00000372779.4 |
PPT1
|
palmitoyl-protein thioesterase 1 |
| chr1_-_183538319 | 0.14 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr19_+_41856816 | 0.14 |
ENST00000539627.1
|
TMEM91
|
transmembrane protein 91 |
| chr9_-_123555655 | 0.14 |
ENST00000340778.5
ENST00000453291.1 ENST00000608872.1 |
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr4_-_69083720 | 0.14 |
ENST00000432593.3
|
TMPRSS11BNL
|
TMPRSS11B N-terminal like |
| chrX_-_30595959 | 0.14 |
ENST00000378962.3
|
CXorf21
|
chromosome X open reading frame 21 |
| chr19_-_48018203 | 0.14 |
ENST00000595227.1
ENST00000593761.1 ENST00000263354.3 |
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr12_-_39734783 | 0.14 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFE2L1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0043311 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.4 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.2 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.1 | 0.3 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.6 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.3 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.3 | GO:1901805 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.2 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 0.3 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 0.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.4 | GO:0031620 | regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) |
| 0.1 | 0.3 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.3 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.4 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) positive regulation of tooth mineralization(GO:0070172) |
| 0.1 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.3 | GO:0042335 | cuticle development(GO:0042335) hypophysis morphogenesis(GO:0048850) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.2 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.8 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.4 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.2 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.3 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.4 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.3 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.3 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:1903979 | negative regulation of microglial cell activation(GO:1903979) |
| 0.0 | 0.1 | GO:0019860 | uracil metabolic process(GO:0019860) intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.5 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.1 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.2 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.3 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.5 | GO:0007099 | centriole replication(GO:0007099) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.3 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 0.2 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.0 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.6 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 0.4 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.1 | 0.5 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.1 | 0.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.5 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.2 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.3 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 0.3 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.3 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.1 | 0.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.4 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0047035 | alcohol dehydrogenase (NAD) activity(GO:0004022) testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.0 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 0.0 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.4 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |