Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
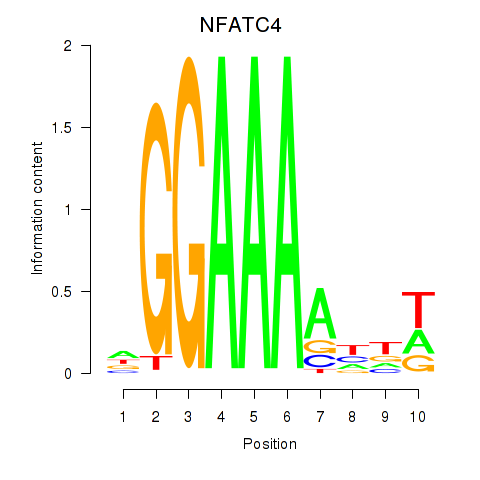
Results for NFATC4
Z-value: 0.88
Transcription factors associated with NFATC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC4
|
ENSG00000100968.9 | nuclear factor of activated T cells 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC4 | hg19_v2_chr14_+_24837226_24837547 | -0.74 | 9.3e-02 | Click! |
Activity profile of NFATC4 motif
Sorted Z-values of NFATC4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_86116396 | 0.51 |
ENST00000455121.3
|
AC105053.4
|
AC105053.4 |
| chr22_-_39548443 | 0.41 |
ENST00000401405.3
|
CBX7
|
chromobox homolog 7 |
| chr12_-_58329888 | 0.39 |
ENST00000546580.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr11_-_126174186 | 0.37 |
ENST00000524964.1
|
RP11-712L6.5
|
Uncharacterized protein |
| chr14_-_51135005 | 0.37 |
ENST00000556735.1
|
SAV1
|
salvador homolog 1 (Drosophila) |
| chrX_+_100224676 | 0.26 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr1_-_100643765 | 0.25 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr2_+_201994208 | 0.25 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr4_+_26322409 | 0.25 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr6_-_53530474 | 0.25 |
ENST00000370905.3
|
KLHL31
|
kelch-like family member 31 |
| chr2_-_165697717 | 0.25 |
ENST00000439313.1
|
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr4_+_175205038 | 0.24 |
ENST00000457424.2
ENST00000514712.1 |
CEP44
|
centrosomal protein 44kDa |
| chr14_+_32546145 | 0.24 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr16_-_18937072 | 0.24 |
ENST00000569122.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr8_-_25281747 | 0.24 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr14_+_75536335 | 0.23 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr3_-_49837254 | 0.23 |
ENST00000412678.2
ENST00000343366.4 ENST00000487256.1 |
CDHR4
|
cadherin-related family member 4 |
| chr11_-_62457371 | 0.21 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr14_+_96858433 | 0.21 |
ENST00000267584.4
|
AK7
|
adenylate kinase 7 |
| chr14_+_21156915 | 0.21 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr14_+_32546274 | 0.21 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr7_+_23210760 | 0.21 |
ENST00000366347.4
|
AC005082.1
|
Uncharacterized protein |
| chr19_+_10959043 | 0.20 |
ENST00000397820.4
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr12_+_51318513 | 0.20 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr8_+_101349823 | 0.20 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr14_-_54955376 | 0.20 |
ENST00000553333.1
|
GMFB
|
glia maturation factor, beta |
| chr1_+_212782012 | 0.19 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
| chr18_-_3247084 | 0.19 |
ENST00000609924.1
|
RP13-270P17.3
|
RP13-270P17.3 |
| chr5_+_63461642 | 0.19 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr2_+_191208196 | 0.19 |
ENST00000392329.2
ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr17_-_34207295 | 0.18 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chrX_-_48827976 | 0.18 |
ENST00000218176.3
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr11_+_100862811 | 0.18 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr1_+_78470530 | 0.18 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr10_-_116444371 | 0.18 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr12_-_47219733 | 0.18 |
ENST00000547477.1
ENST00000447411.1 ENST00000266579.4 |
SLC38A4
|
solute carrier family 38, member 4 |
| chr15_-_77712477 | 0.17 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr2_+_201994042 | 0.17 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr7_-_152373216 | 0.17 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr11_+_62475130 | 0.17 |
ENST00000294117.5
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr17_-_58603482 | 0.17 |
ENST00000585368.1
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr14_+_58754751 | 0.17 |
ENST00000598233.1
|
AL132989.1
|
AL132989.1 |
| chr2_+_98703595 | 0.17 |
ENST00000435344.1
|
VWA3B
|
von Willebrand factor A domain containing 3B |
| chr1_-_113161730 | 0.17 |
ENST00000544629.1
ENST00000543570.1 ENST00000360743.4 ENST00000490067.1 ENST00000343210.7 ENST00000369666.1 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr10_-_16563870 | 0.16 |
ENST00000298943.3
|
C1QL3
|
complement component 1, q subcomponent-like 3 |
| chr17_-_58499766 | 0.16 |
ENST00000588898.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr5_-_43412418 | 0.16 |
ENST00000537013.1
ENST00000361115.4 |
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr11_-_124806297 | 0.16 |
ENST00000298251.4
|
HEPACAM
|
hepatic and glial cell adhesion molecule |
| chr4_+_175205100 | 0.16 |
ENST00000515299.1
|
CEP44
|
centrosomal protein 44kDa |
| chr2_+_191208601 | 0.16 |
ENST00000413239.1
ENST00000431594.1 ENST00000444194.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr17_+_67590125 | 0.16 |
ENST00000591334.1
|
AC003051.1
|
AC003051.1 |
| chr3_+_98451532 | 0.15 |
ENST00000486334.2
ENST00000394162.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr6_-_146057144 | 0.15 |
ENST00000367519.3
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr15_+_84841242 | 0.15 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr2_-_207629997 | 0.15 |
ENST00000454776.2
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr17_-_1613663 | 0.15 |
ENST00000330676.6
|
TLCD2
|
TLC domain containing 2 |
| chr2_+_98703695 | 0.15 |
ENST00000451075.2
|
VWA3B
|
von Willebrand factor A domain containing 3B |
| chr1_-_151826173 | 0.15 |
ENST00000368817.5
|
THEM5
|
thioesterase superfamily member 5 |
| chr11_-_46142948 | 0.15 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr17_+_44701402 | 0.15 |
ENST00000575068.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr1_-_145470383 | 0.15 |
ENST00000369314.1
ENST00000369313.3 |
POLR3GL
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)-like |
| chr9_-_13165457 | 0.14 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr13_+_102104980 | 0.14 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr8_+_28747884 | 0.14 |
ENST00000287701.10
ENST00000444075.1 ENST00000403668.2 ENST00000519662.1 ENST00000558662.1 ENST00000523613.1 ENST00000560599.1 ENST00000397358.3 |
HMBOX1
|
homeobox containing 1 |
| chrX_+_22056165 | 0.14 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr12_-_49075941 | 0.14 |
ENST00000553086.1
ENST00000548304.1 |
KANSL2
|
KAT8 regulatory NSL complex subunit 2 |
| chr11_-_5537920 | 0.13 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr6_-_26235206 | 0.13 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr11_-_61735029 | 0.13 |
ENST00000526640.1
|
FTH1
|
ferritin, heavy polypeptide 1 |
| chr1_+_99127225 | 0.13 |
ENST00000370189.5
ENST00000529992.1 |
SNX7
|
sorting nexin 7 |
| chr8_+_70404996 | 0.13 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr1_+_99127265 | 0.13 |
ENST00000306121.3
|
SNX7
|
sorting nexin 7 |
| chr2_+_191208791 | 0.13 |
ENST00000423767.1
ENST00000451089.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr3_+_178865887 | 0.13 |
ENST00000477735.1
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr2_+_191208656 | 0.13 |
ENST00000458647.1
|
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr7_-_105319536 | 0.13 |
ENST00000477775.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr14_-_30396948 | 0.13 |
ENST00000331968.5
|
PRKD1
|
protein kinase D1 |
| chr22_-_29107919 | 0.13 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr3_+_154797781 | 0.13 |
ENST00000382989.3
|
MME
|
membrane metallo-endopeptidase |
| chr3_+_138340067 | 0.13 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr17_+_6900201 | 0.12 |
ENST00000480801.1
|
ALOX12
|
arachidonate 12-lipoxygenase |
| chr11_-_46142615 | 0.12 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr1_-_113162040 | 0.12 |
ENST00000358039.4
ENST00000369668.2 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr5_+_72112418 | 0.12 |
ENST00000454282.1
|
TNPO1
|
transportin 1 |
| chr12_-_95467267 | 0.12 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr5_+_15500280 | 0.12 |
ENST00000504595.1
|
FBXL7
|
F-box and leucine-rich repeat protein 7 |
| chr17_+_65375082 | 0.12 |
ENST00000584471.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chrX_+_23928500 | 0.12 |
ENST00000435707.1
|
CXorf58
|
chromosome X open reading frame 58 |
| chr20_+_33464238 | 0.12 |
ENST00000360596.2
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr11_-_46113756 | 0.12 |
ENST00000531959.1
|
PHF21A
|
PHD finger protein 21A |
| chr12_+_56511943 | 0.12 |
ENST00000257940.2
ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10
ESYT1
|
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr21_+_17791838 | 0.12 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_-_234667504 | 0.12 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr18_+_3451584 | 0.12 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr3_-_93747425 | 0.12 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr17_+_40996590 | 0.12 |
ENST00000253799.3
ENST00000452774.2 |
AOC2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr14_+_54863682 | 0.12 |
ENST00000543789.2
ENST00000442975.2 ENST00000458126.2 ENST00000556102.2 |
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr6_-_131384412 | 0.12 |
ENST00000445890.2
ENST00000368128.2 ENST00000337057.3 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr1_-_155881156 | 0.12 |
ENST00000539040.1
ENST00000368323.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr10_+_25305524 | 0.12 |
ENST00000524413.1
ENST00000376356.4 |
THNSL1
|
threonine synthase-like 1 (S. cerevisiae) |
| chr1_-_161337662 | 0.12 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr15_-_90437195 | 0.12 |
ENST00000560940.1
ENST00000558011.1 |
AP3S2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr12_+_41086215 | 0.11 |
ENST00000547702.1
ENST00000551424.1 |
CNTN1
|
contactin 1 |
| chr17_+_5389605 | 0.11 |
ENST00000576988.1
ENST00000576570.1 ENST00000573759.1 |
MIS12
|
MIS12 kinetochore complex component |
| chr19_+_37095719 | 0.11 |
ENST00000423582.1
|
ZNF382
|
zinc finger protein 382 |
| chr1_-_68698197 | 0.11 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr12_-_54653313 | 0.11 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr5_+_158690089 | 0.11 |
ENST00000296786.6
|
UBLCP1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr7_-_64023441 | 0.11 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr5_+_102090974 | 0.11 |
ENST00000512073.1
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chrX_+_129473916 | 0.11 |
ENST00000545805.1
ENST00000543953.1 ENST00000218197.5 |
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr2_+_204193101 | 0.11 |
ENST00000430418.1
ENST00000424558.1 ENST00000261016.6 |
ABI2
|
abl-interactor 2 |
| chr2_+_98703643 | 0.11 |
ENST00000477737.1
|
VWA3B
|
von Willebrand factor A domain containing 3B |
| chr6_-_131384373 | 0.11 |
ENST00000392427.3
ENST00000525271.1 ENST00000527411.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr3_-_197024394 | 0.11 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr3_+_160559931 | 0.11 |
ENST00000464260.1
ENST00000295839.9 |
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chrX_+_100663243 | 0.11 |
ENST00000316594.5
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr2_-_97405775 | 0.11 |
ENST00000264963.4
ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L
|
lectin, mannose-binding 2-like |
| chr1_-_150780757 | 0.11 |
ENST00000271651.3
|
CTSK
|
cathepsin K |
| chr3_-_46068969 | 0.11 |
ENST00000542109.1
ENST00000395946.2 |
XCR1
|
chemokine (C motif) receptor 1 |
| chr5_+_154238149 | 0.11 |
ENST00000519430.1
ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr4_-_48683188 | 0.11 |
ENST00000505759.1
|
FRYL
|
FRY-like |
| chr4_+_26322185 | 0.11 |
ENST00000361572.6
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr21_+_17791648 | 0.11 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr2_-_175113301 | 0.11 |
ENST00000344357.5
ENST00000284719.3 |
OLA1
|
Obg-like ATPase 1 |
| chr17_-_44657017 | 0.11 |
ENST00000573185.1
ENST00000570550.1 ENST00000445552.2 ENST00000336125.5 ENST00000329240.4 ENST00000337845.7 |
ARL17A
|
ADP-ribosylation factor-like 17A |
| chr22_-_39548511 | 0.11 |
ENST00000434260.1
|
CBX7
|
chromobox homolog 7 |
| chr14_+_81421921 | 0.10 |
ENST00000554263.1
ENST00000554435.1 |
TSHR
|
thyroid stimulating hormone receptor |
| chr12_+_64798826 | 0.10 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr12_-_39734783 | 0.10 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr14_+_54863667 | 0.10 |
ENST00000335183.6
|
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr18_-_64271316 | 0.10 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr2_-_207630033 | 0.10 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr4_+_175204818 | 0.10 |
ENST00000503780.1
|
CEP44
|
centrosomal protein 44kDa |
| chr14_-_61748550 | 0.10 |
ENST00000555868.1
|
TMEM30B
|
transmembrane protein 30B |
| chr6_-_132834184 | 0.10 |
ENST00000367941.2
ENST00000367937.4 |
STX7
|
syntaxin 7 |
| chr11_-_46142505 | 0.10 |
ENST00000524497.1
ENST00000418153.2 |
PHF21A
|
PHD finger protein 21A |
| chr6_-_159065741 | 0.10 |
ENST00000367085.3
ENST00000367089.3 |
DYNLT1
|
dynein, light chain, Tctex-type 1 |
| chr12_+_21654714 | 0.10 |
ENST00000542038.1
ENST00000540141.1 ENST00000229314.5 |
GOLT1B
|
golgi transport 1B |
| chr10_-_7453445 | 0.10 |
ENST00000379713.3
ENST00000397167.1 ENST00000397160.3 |
SFMBT2
|
Scm-like with four mbt domains 2 |
| chr1_-_8075693 | 0.10 |
ENST00000467067.1
|
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr2_+_29353520 | 0.10 |
ENST00000438819.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr2_-_55277692 | 0.10 |
ENST00000394611.2
|
RTN4
|
reticulon 4 |
| chr1_+_206317591 | 0.10 |
ENST00000432969.2
|
CTSE
|
cathepsin E |
| chr11_-_47600320 | 0.10 |
ENST00000525720.1
ENST00000531067.1 ENST00000533290.1 ENST00000529499.1 ENST00000529946.1 ENST00000526005.1 ENST00000395288.2 ENST00000534239.1 |
KBTBD4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr17_+_52978185 | 0.10 |
ENST00000572405.1
ENST00000572158.1 ENST00000540336.1 ENST00000572298.1 ENST00000536554.1 ENST00000575333.1 ENST00000570499.1 ENST00000572576.1 |
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr2_-_191115229 | 0.10 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr9_+_26956371 | 0.10 |
ENST00000380062.5
ENST00000518614.1 |
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr6_+_122720681 | 0.10 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr5_-_42812143 | 0.10 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr7_-_107204337 | 0.10 |
ENST00000605888.1
ENST00000347053.3 |
COG5
|
component of oligomeric golgi complex 5 |
| chr3_+_98451093 | 0.10 |
ENST00000483910.1
ENST00000460774.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr18_+_5238549 | 0.10 |
ENST00000580684.1
|
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr12_-_56105880 | 0.10 |
ENST00000557257.1
|
ITGA7
|
integrin, alpha 7 |
| chr3_+_142342228 | 0.10 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr12_-_76461249 | 0.10 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr10_+_114710516 | 0.09 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr15_-_77712429 | 0.09 |
ENST00000564328.1
ENST00000558305.1 |
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr11_+_1942580 | 0.09 |
ENST00000381558.1
|
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr12_+_3600356 | 0.09 |
ENST00000382622.3
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr12_-_21910775 | 0.09 |
ENST00000539782.1
|
LDHB
|
lactate dehydrogenase B |
| chr4_+_89300158 | 0.09 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr20_+_35201993 | 0.09 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr3_+_69812877 | 0.09 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr1_+_65613513 | 0.09 |
ENST00000395334.2
|
AK4
|
adenylate kinase 4 |
| chr11_-_47600549 | 0.09 |
ENST00000430070.2
|
KBTBD4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr3_+_63953415 | 0.09 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr7_-_99679324 | 0.09 |
ENST00000292393.5
ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3
|
zinc finger protein 3 |
| chr6_-_56716686 | 0.09 |
ENST00000520645.1
|
DST
|
dystonin |
| chr11_+_114271367 | 0.09 |
ENST00000544582.1
ENST00000545678.1 |
RBM7
|
RNA binding motif protein 7 |
| chr15_-_72563585 | 0.09 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr1_-_52521831 | 0.09 |
ENST00000371626.4
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr4_-_175204765 | 0.09 |
ENST00000513696.1
ENST00000503293.1 |
FBXO8
|
F-box protein 8 |
| chr4_-_176733897 | 0.09 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr1_+_155178518 | 0.09 |
ENST00000316721.4
|
MTX1
|
metaxin 1 |
| chrX_-_102943022 | 0.09 |
ENST00000433176.2
|
MORF4L2
|
mortality factor 4 like 2 |
| chr12_-_47226152 | 0.09 |
ENST00000546940.1
|
SLC38A4
|
solute carrier family 38, member 4 |
| chr4_-_52883786 | 0.09 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr3_-_42642560 | 0.09 |
ENST00000417572.1
|
SEC22C
|
SEC22 vesicle trafficking protein homolog C (S. cerevisiae) |
| chrX_-_84634708 | 0.09 |
ENST00000373145.3
|
POF1B
|
premature ovarian failure, 1B |
| chr6_-_31510181 | 0.09 |
ENST00000458640.1
ENST00000396172.1 ENST00000417556.2 |
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr5_+_68462944 | 0.09 |
ENST00000506572.1
|
CCNB1
|
cyclin B1 |
| chrX_+_70503526 | 0.09 |
ENST00000413858.1
ENST00000450092.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr10_-_62704005 | 0.08 |
ENST00000337910.5
|
RHOBTB1
|
Rho-related BTB domain containing 1 |
| chr7_-_92157747 | 0.08 |
ENST00000428214.1
ENST00000438045.1 |
PEX1
|
peroxisomal biogenesis factor 1 |
| chr12_+_10460417 | 0.08 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr7_-_92777606 | 0.08 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr19_+_52772832 | 0.08 |
ENST00000593703.1
ENST00000601711.1 ENST00000599581.1 |
ZNF766
|
zinc finger protein 766 |
| chr8_+_94767109 | 0.08 |
ENST00000409623.3
ENST00000453906.1 ENST00000521517.1 |
TMEM67
|
transmembrane protein 67 |
| chr3_+_98451275 | 0.08 |
ENST00000265261.6
ENST00000497008.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr16_-_85969774 | 0.08 |
ENST00000598933.1
|
RP11-542M13.3
|
RP11-542M13.3 |
| chr6_-_131384347 | 0.08 |
ENST00000530481.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr1_-_155880672 | 0.08 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr1_+_40627038 | 0.08 |
ENST00000372771.4
|
RLF
|
rearranged L-myc fusion |
| chr1_-_150207017 | 0.08 |
ENST00000369119.3
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr2_+_223916862 | 0.08 |
ENST00000604125.1
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4 |
| chr3_-_149375783 | 0.08 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr11_+_65190245 | 0.08 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr2_+_27301435 | 0.08 |
ENST00000380320.4
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr1_-_151299842 | 0.08 |
ENST00000438243.2
ENST00000489223.2 ENST00000368873.1 ENST00000430800.1 ENST00000368872.1 |
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0046883 | hormone secretion(GO:0046879) regulation of hormone secretion(GO:0046883) |
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.4 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.0 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0097018 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.4 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:2001302 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.1 | GO:2000775 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0038194 | thyroid-stimulating hormone signaling pathway(GO:0038194) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) protein kinase D signaling(GO:0089700) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.0 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.4 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.2 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.0 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.0 | 0.1 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.0 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.1 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.0 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.1 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.0 | GO:0070858 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) negative regulation of bile acid biosynthetic process(GO:0070858) acinar cell differentiation(GO:0090425) negative regulation of bile acid metabolic process(GO:1904252) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:1903764 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.2 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.1 | 0.3 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.1 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.0 | 0.0 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.0 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |