Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
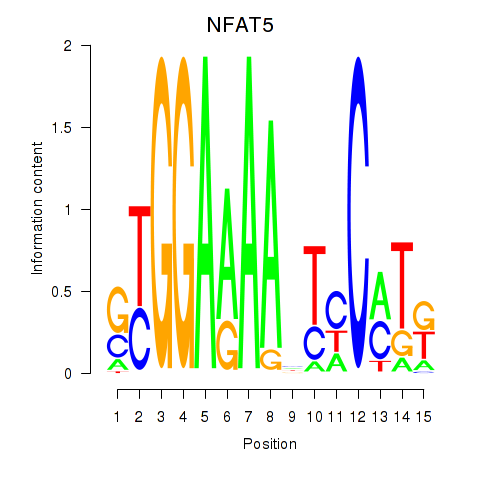
Results for NFAT5
Z-value: 0.73
Transcription factors associated with NFAT5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFAT5
|
ENSG00000102908.16 | nuclear factor of activated T cells 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFAT5 | hg19_v2_chr16_+_69599861_69599887 | -0.77 | 7.6e-02 | Click! |
Activity profile of NFAT5 motif
Sorted Z-values of NFAT5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_103589789 | 0.55 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr6_+_138188351 | 0.51 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr14_+_95078714 | 0.49 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr12_+_26274917 | 0.43 |
ENST00000538142.1
|
SSPN
|
sarcospan |
| chr21_+_17792672 | 0.36 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_95975672 | 0.33 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr4_+_105828492 | 0.31 |
ENST00000506148.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr2_+_47799601 | 0.30 |
ENST00000601243.1
|
AC138655.1
|
CDNA: FLJ23120 fis, clone LNG07989; HCG1987724; Uncharacterized protein |
| chr6_+_138188551 | 0.29 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr10_+_135122906 | 0.28 |
ENST00000368554.4
|
ZNF511
|
zinc finger protein 511 |
| chr2_-_20101385 | 0.27 |
ENST00000431392.1
|
TTC32
|
tetratricopeptide repeat domain 32 |
| chr12_+_46777450 | 0.25 |
ENST00000551503.1
|
RP11-96H19.1
|
RP11-96H19.1 |
| chr14_+_97059070 | 0.24 |
ENST00000553378.1
ENST00000555496.1 |
RP11-433J8.1
|
RP11-433J8.1 |
| chr13_-_30160925 | 0.24 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr12_+_120502441 | 0.23 |
ENST00000446727.2
|
CCDC64
|
coiled-coil domain containing 64 |
| chr12_-_78934441 | 0.22 |
ENST00000546865.1
ENST00000547089.1 |
RP11-171L9.1
|
RP11-171L9.1 |
| chr19_+_45254529 | 0.22 |
ENST00000444487.1
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr14_-_25479811 | 0.21 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr16_+_89988259 | 0.21 |
ENST00000554444.1
ENST00000556565.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr3_-_42003613 | 0.20 |
ENST00000414606.1
|
ULK4
|
unc-51 like kinase 4 |
| chr3_+_122399697 | 0.20 |
ENST00000494811.1
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr17_+_45908974 | 0.20 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr19_-_51017127 | 0.19 |
ENST00000389208.4
|
ASPDH
|
aspartate dehydrogenase domain containing |
| chr19_+_17905594 | 0.19 |
ENST00000599265.1
|
B3GNT3
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
| chr15_-_49170219 | 0.19 |
ENST00000558220.1
ENST00000537958.1 |
SHC4
|
SHC (Src homology 2 domain containing) family, member 4 |
| chr4_+_17579110 | 0.19 |
ENST00000606142.1
|
LAP3
|
leucine aminopeptidase 3 |
| chr3_-_9834463 | 0.18 |
ENST00000439043.1
|
TADA3
|
transcriptional adaptor 3 |
| chr17_+_6915730 | 0.18 |
ENST00000548577.1
|
RNASEK
|
ribonuclease, RNase K |
| chr13_-_30996373 | 0.17 |
ENST00000420694.1
|
LINC01058
|
long intergenic non-protein coding RNA 1058 |
| chr15_+_100348193 | 0.16 |
ENST00000558188.1
|
CTD-2054N24.2
|
Uncharacterized protein |
| chr7_+_141478242 | 0.16 |
ENST00000247881.2
|
TAS2R4
|
taste receptor, type 2, member 4 |
| chrX_+_48755183 | 0.16 |
ENST00000376563.1
ENST00000376566.4 |
PQBP1
|
polyglutamine binding protein 1 |
| chr17_+_38121772 | 0.16 |
ENST00000577447.1
|
GSDMA
|
gasdermin A |
| chr11_-_75062829 | 0.16 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr17_-_39093672 | 0.15 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr4_+_105828537 | 0.15 |
ENST00000515649.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr3_+_9834758 | 0.15 |
ENST00000485273.1
ENST00000433034.1 ENST00000397256.1 |
ARPC4
ARPC4-TTLL3
|
actin related protein 2/3 complex, subunit 4, 20kDa ARPC4-TTLL3 readthrough |
| chr3_+_184079492 | 0.15 |
ENST00000456318.1
ENST00000412877.1 ENST00000438240.1 |
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr6_+_138188378 | 0.15 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr3_+_77088989 | 0.14 |
ENST00000461745.1
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr3_+_9834179 | 0.14 |
ENST00000498623.2
|
ARPC4
|
actin related protein 2/3 complex, subunit 4, 20kDa |
| chr1_-_246670614 | 0.14 |
ENST00000403792.3
|
SMYD3
|
SET and MYND domain containing 3 |
| chr4_-_83483360 | 0.14 |
ENST00000449862.2
|
TMEM150C
|
transmembrane protein 150C |
| chr6_-_134495992 | 0.14 |
ENST00000475719.2
ENST00000367857.5 ENST00000237305.7 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr17_+_18684468 | 0.13 |
ENST00000574226.1
ENST00000575261.1 |
TVP23B
|
trans-golgi network vesicle protein 23 homolog B (S. cerevisiae) |
| chr19_+_44037546 | 0.13 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr12_+_22778291 | 0.13 |
ENST00000545979.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr7_+_65670186 | 0.13 |
ENST00000304842.5
ENST00000442120.1 |
TPST1
|
tyrosylprotein sulfotransferase 1 |
| chr17_+_4853442 | 0.12 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chrX_+_135579670 | 0.12 |
ENST00000218364.4
|
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr4_+_17578815 | 0.12 |
ENST00000226299.4
|
LAP3
|
leucine aminopeptidase 3 |
| chr6_+_12007897 | 0.12 |
ENST00000437559.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr5_+_140186647 | 0.12 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr1_+_18958008 | 0.12 |
ENST00000420770.2
ENST00000400661.3 |
PAX7
|
paired box 7 |
| chr3_+_177159744 | 0.11 |
ENST00000439009.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr8_+_96037205 | 0.11 |
ENST00000396124.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr3_-_9834375 | 0.11 |
ENST00000343450.2
ENST00000301964.2 |
TADA3
|
transcriptional adaptor 3 |
| chr18_-_33702078 | 0.11 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr3_+_9834227 | 0.11 |
ENST00000287613.7
ENST00000397261.3 |
ARPC4
|
actin related protein 2/3 complex, subunit 4, 20kDa |
| chr14_+_100594914 | 0.11 |
ENST00000554695.1
|
EVL
|
Enah/Vasp-like |
| chr5_-_96209315 | 0.11 |
ENST00000504056.1
|
CTD-2260A17.2
|
Uncharacterized protein |
| chr11_-_2924970 | 0.11 |
ENST00000533594.1
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr17_+_68071389 | 0.11 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr3_+_47866490 | 0.10 |
ENST00000457607.1
|
DHX30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr3_+_25469724 | 0.10 |
ENST00000437042.2
|
RARB
|
retinoic acid receptor, beta |
| chr13_+_113030658 | 0.10 |
ENST00000414180.1
ENST00000443541.1 |
SPACA7
|
sperm acrosome associated 7 |
| chr16_+_2079501 | 0.10 |
ENST00000563587.1
|
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr16_-_67514982 | 0.10 |
ENST00000565835.1
ENST00000540149.1 ENST00000290949.3 |
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr19_-_46706186 | 0.10 |
ENST00000601263.1
ENST00000598754.1 ENST00000594027.1 ENST00000598518.1 ENST00000599817.1 ENST00000595673.1 |
AC006262.5
|
AC006262.5 |
| chr5_-_150521192 | 0.10 |
ENST00000523714.1
ENST00000521749.1 |
ANXA6
|
annexin A6 |
| chr12_-_54813229 | 0.10 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr3_-_48594248 | 0.10 |
ENST00000545984.1
ENST00000232375.3 ENST00000416568.1 ENST00000383734.2 ENST00000541519.1 ENST00000412035.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr1_-_246670519 | 0.10 |
ENST00000388985.4
ENST00000490107.1 |
SMYD3
|
SET and MYND domain containing 3 |
| chr4_-_83483395 | 0.10 |
ENST00000515780.2
|
TMEM150C
|
transmembrane protein 150C |
| chr3_+_8543393 | 0.10 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr1_+_201592013 | 0.10 |
ENST00000593583.1
|
AC096677.1
|
Uncharacterized protein ENSP00000471857 |
| chr11_-_75062730 | 0.10 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr10_-_135122603 | 0.10 |
ENST00000368563.2
|
TUBGCP2
|
tubulin, gamma complex associated protein 2 |
| chr17_-_46608272 | 0.09 |
ENST00000577092.1
ENST00000239174.6 |
HOXB1
|
homeobox B1 |
| chr1_-_145470383 | 0.09 |
ENST00000369314.1
ENST00000369313.3 |
POLR3GL
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)-like |
| chr15_-_102285007 | 0.09 |
ENST00000560292.2
|
RP11-89K11.1
|
Uncharacterized protein |
| chr7_-_100888337 | 0.09 |
ENST00000223136.4
|
FIS1
|
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) |
| chr3_-_47950745 | 0.09 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr3_+_127770455 | 0.09 |
ENST00000464451.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr11_+_65154070 | 0.09 |
ENST00000317568.5
ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8
|
FERM domain containing 8 |
| chr11_+_131240593 | 0.09 |
ENST00000539799.1
|
NTM
|
neurotrimin |
| chr6_-_31926208 | 0.08 |
ENST00000454913.1
ENST00000436289.2 |
NELFE
|
negative elongation factor complex member E |
| chr17_+_68071458 | 0.08 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr6_-_131291572 | 0.08 |
ENST00000529208.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr6_+_116832789 | 0.08 |
ENST00000368599.3
|
FAM26E
|
family with sequence similarity 26, member E |
| chr13_-_67804445 | 0.08 |
ENST00000456367.1
ENST00000377861.3 ENST00000544246.1 |
PCDH9
|
protocadherin 9 |
| chr1_-_85870177 | 0.08 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr12_+_10365082 | 0.08 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr12_+_101988627 | 0.08 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr1_+_155006300 | 0.08 |
ENST00000295542.1
ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr10_-_124639062 | 0.08 |
ENST00000368898.3
ENST00000368896.1 ENST00000545804.1 |
FAM24B
CUZD1
|
family with sequence similarity 24, member B CUB and zona pellucida-like domains 1 |
| chr3_+_49027308 | 0.07 |
ENST00000383729.4
ENST00000343546.4 |
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr15_+_54901540 | 0.07 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr6_+_4890226 | 0.07 |
ENST00000343762.5
|
CDYL
|
chromodomain protein, Y-like |
| chr11_+_60145948 | 0.07 |
ENST00000300184.3
ENST00000358246.1 |
MS4A7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr7_+_95401851 | 0.07 |
ENST00000447467.2
|
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr12_+_101988774 | 0.07 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr4_-_74847800 | 0.07 |
ENST00000296029.3
|
PF4
|
platelet factor 4 |
| chrX_-_128977875 | 0.07 |
ENST00000406492.2
|
ZDHHC9
|
zinc finger, DHHC-type containing 9 |
| chr5_-_137090028 | 0.07 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr15_-_75134992 | 0.07 |
ENST00000568667.1
|
ULK3
|
unc-51 like kinase 3 |
| chr14_+_100848311 | 0.07 |
ENST00000542471.2
|
WDR25
|
WD repeat domain 25 |
| chr4_-_76861392 | 0.07 |
ENST00000505594.1
|
NAAA
|
N-acylethanolamine acid amidase |
| chr7_-_92855762 | 0.07 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr17_+_6658878 | 0.07 |
ENST00000574394.1
|
XAF1
|
XIAP associated factor 1 |
| chr16_-_85969774 | 0.07 |
ENST00000598933.1
|
RP11-542M13.3
|
RP11-542M13.3 |
| chr17_+_17991197 | 0.07 |
ENST00000225729.3
|
DRG2
|
developmentally regulated GTP binding protein 2 |
| chr2_+_108994466 | 0.07 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr12_-_108991778 | 0.06 |
ENST00000549447.1
|
TMEM119
|
transmembrane protein 119 |
| chr6_+_44094627 | 0.06 |
ENST00000259746.9
|
TMEM63B
|
transmembrane protein 63B |
| chr7_+_1127723 | 0.06 |
ENST00000397088.3
|
GPER1
|
G protein-coupled estrogen receptor 1 |
| chr6_+_130339710 | 0.06 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr1_+_111889212 | 0.06 |
ENST00000369737.4
|
PIFO
|
primary cilia formation |
| chr16_+_2079637 | 0.06 |
ENST00000561844.1
|
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chrX_+_18725758 | 0.06 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr7_+_1126461 | 0.06 |
ENST00000297469.3
|
GPER1
|
G protein-coupled estrogen receptor 1 |
| chr19_+_49999631 | 0.06 |
ENST00000270625.2
ENST00000596873.1 ENST00000594493.1 ENST00000599561.1 |
RPS11
|
ribosomal protein S11 |
| chr12_+_53848549 | 0.06 |
ENST00000439930.3
ENST00000548933.1 ENST00000562264.1 |
PCBP2
|
poly(rC) binding protein 2 |
| chr6_+_152011628 | 0.06 |
ENST00000404742.1
ENST00000440973.1 |
ESR1
|
estrogen receptor 1 |
| chr1_-_155006254 | 0.06 |
ENST00000295536.5
|
DCST2
|
DC-STAMP domain containing 2 |
| chr18_+_11103 | 0.06 |
ENST00000575820.1
ENST00000572573.1 |
AP005530.1
|
AP005530.1 |
| chr14_+_91580357 | 0.06 |
ENST00000298858.4
ENST00000521081.1 ENST00000520328.1 ENST00000256324.10 ENST00000524232.1 ENST00000522170.1 ENST00000519950.1 ENST00000523879.1 ENST00000521077.2 ENST00000518665.2 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr17_-_202579 | 0.06 |
ENST00000577079.1
ENST00000331302.7 ENST00000536489.2 |
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr19_+_17420340 | 0.06 |
ENST00000359866.4
|
DDA1
|
DET1 and DDB1 associated 1 |
| chr7_-_100888313 | 0.06 |
ENST00000442303.1
|
FIS1
|
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) |
| chr1_-_115300592 | 0.06 |
ENST00000261443.5
ENST00000534699.1 ENST00000339438.6 ENST00000529046.1 ENST00000525970.1 ENST00000369530.1 ENST00000530886.1 |
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr14_+_39644425 | 0.06 |
ENST00000556530.1
|
PNN
|
pinin, desmosome associated protein |
| chr2_+_48796120 | 0.06 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr12_+_110011571 | 0.06 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr11_-_67120974 | 0.06 |
ENST00000539074.1
ENST00000312419.3 |
POLD4
|
polymerase (DNA-directed), delta 4, accessory subunit |
| chr2_-_223163465 | 0.06 |
ENST00000409828.3
ENST00000350526.4 ENST00000258387.5 ENST00000409551.3 ENST00000392069.2 ENST00000344493.4 ENST00000336840.6 ENST00000392070.2 |
PAX3
|
paired box 3 |
| chr2_-_242211359 | 0.05 |
ENST00000444092.1
|
HDLBP
|
high density lipoprotein binding protein |
| chr6_+_30103885 | 0.05 |
ENST00000396581.1
|
TRIM40
|
tripartite motif containing 40 |
| chr7_+_1126437 | 0.05 |
ENST00000413368.1
ENST00000397092.1 |
GPER1
|
G protein-coupled estrogen receptor 1 |
| chr19_+_14142535 | 0.05 |
ENST00000263379.2
|
IL27RA
|
interleukin 27 receptor, alpha |
| chr15_+_43885799 | 0.05 |
ENST00000449946.1
ENST00000417289.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr7_+_128399002 | 0.05 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr3_+_8543561 | 0.05 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr1_-_57045228 | 0.05 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr7_-_102119342 | 0.05 |
ENST00000393794.3
ENST00000292614.5 |
POLR2J
|
polymerase (RNA) II (DNA directed) polypeptide J, 13.3kDa |
| chr5_+_140261703 | 0.05 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr19_+_42212501 | 0.05 |
ENST00000398599.4
|
CEACAM5
|
carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr19_+_859425 | 0.05 |
ENST00000327726.6
|
CFD
|
complement factor D (adipsin) |
| chr3_-_49131013 | 0.05 |
ENST00000424300.1
|
QRICH1
|
glutamine-rich 1 |
| chr18_-_52989217 | 0.05 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr1_-_155006224 | 0.05 |
ENST00000368424.3
|
DCST2
|
DC-STAMP domain containing 2 |
| chr6_+_73331814 | 0.05 |
ENST00000370392.1
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr7_+_149570049 | 0.05 |
ENST00000421974.2
ENST00000456496.2 |
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2 |
| chr4_-_155511887 | 0.05 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr12_+_10365404 | 0.05 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr11_-_18063973 | 0.05 |
ENST00000528338.1
|
TPH1
|
tryptophan hydroxylase 1 |
| chr17_+_40950900 | 0.05 |
ENST00000588527.1
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr11_+_131781290 | 0.05 |
ENST00000425719.2
ENST00000374784.1 |
NTM
|
neurotrimin |
| chr16_-_51185172 | 0.05 |
ENST00000251020.4
|
SALL1
|
spalt-like transcription factor 1 |
| chr19_-_44100275 | 0.04 |
ENST00000422989.1
ENST00000598324.1 |
IRGQ
|
immunity-related GTPase family, Q |
| chr10_-_69455873 | 0.04 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr17_+_9745786 | 0.04 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr1_-_169599314 | 0.04 |
ENST00000367786.2
ENST00000458599.2 ENST00000367795.2 ENST00000263686.6 |
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr19_+_48828788 | 0.04 |
ENST00000594198.1
ENST00000597279.1 ENST00000593437.1 |
EMP3
|
epithelial membrane protein 3 |
| chr11_+_119039069 | 0.04 |
ENST00000422249.1
|
NLRX1
|
NLR family member X1 |
| chr12_+_119772502 | 0.04 |
ENST00000536742.1
ENST00000327554.2 |
CCDC60
|
coiled-coil domain containing 60 |
| chr2_-_238305397 | 0.04 |
ENST00000409809.1
|
COL6A3
|
collagen, type VI, alpha 3 |
| chr7_+_30174574 | 0.04 |
ENST00000409688.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr7_-_99698338 | 0.04 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr14_-_30396948 | 0.04 |
ENST00000331968.5
|
PRKD1
|
protein kinase D1 |
| chr15_+_45028520 | 0.04 |
ENST00000329464.4
|
TRIM69
|
tripartite motif containing 69 |
| chr8_-_128231299 | 0.04 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr2_-_161056762 | 0.04 |
ENST00000428609.2
ENST00000409967.2 |
ITGB6
|
integrin, beta 6 |
| chr2_+_143635067 | 0.04 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr20_-_43753104 | 0.04 |
ENST00000372785.3
|
WFDC12
|
WAP four-disulfide core domain 12 |
| chr10_-_97175444 | 0.04 |
ENST00000486141.2
|
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr1_-_169599353 | 0.04 |
ENST00000367793.2
ENST00000367794.2 ENST00000367792.2 ENST00000367791.2 ENST00000367788.2 |
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr7_+_70229899 | 0.04 |
ENST00000443672.1
|
AUTS2
|
autism susceptibility candidate 2 |
| chrX_-_153583257 | 0.03 |
ENST00000438732.1
|
FLNA
|
filamin A, alpha |
| chr12_+_53848505 | 0.03 |
ENST00000552819.1
ENST00000455667.3 |
PCBP2
|
poly(rC) binding protein 2 |
| chr1_-_201346761 | 0.03 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr17_-_1613663 | 0.03 |
ENST00000330676.6
|
TLCD2
|
TLC domain containing 2 |
| chr4_+_69313145 | 0.03 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr3_+_63805017 | 0.03 |
ENST00000295896.8
|
C3orf49
|
chromosome 3 open reading frame 49 |
| chr7_+_95401877 | 0.03 |
ENST00000524053.1
ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr20_-_23731569 | 0.03 |
ENST00000304749.2
|
CST1
|
cystatin SN |
| chr2_+_238877424 | 0.03 |
ENST00000434655.1
|
UBE2F
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr17_-_40950698 | 0.03 |
ENST00000328434.7
|
COA3
|
cytochrome c oxidase assembly factor 3 |
| chr6_+_153071925 | 0.03 |
ENST00000367244.3
ENST00000367243.3 |
VIP
|
vasoactive intestinal peptide |
| chr1_+_111888890 | 0.03 |
ENST00000369738.4
|
PIFO
|
primary cilia formation |
| chr17_+_38083977 | 0.03 |
ENST00000578802.1
ENST00000578478.1 ENST00000582263.1 |
RP11-387H17.4
|
RP11-387H17.4 |
| chr11_+_119039414 | 0.02 |
ENST00000409991.1
ENST00000292199.2 ENST00000409265.4 ENST00000409109.1 |
NLRX1
|
NLR family member X1 |
| chr1_-_204436344 | 0.02 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr5_-_35230771 | 0.02 |
ENST00000342362.5
|
PRLR
|
prolactin receptor |
| chrX_+_70752917 | 0.02 |
ENST00000373719.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr9_+_70856899 | 0.02 |
ENST00000377342.5
ENST00000478048.1 |
CBWD3
|
COBW domain containing 3 |
| chr12_-_82152444 | 0.02 |
ENST00000549325.1
ENST00000550584.2 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr11_+_73359719 | 0.02 |
ENST00000535129.1
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr15_+_45028753 | 0.02 |
ENST00000338264.4
|
TRIM69
|
tripartite motif containing 69 |
| chr1_-_16345245 | 0.02 |
ENST00000311890.9
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular) |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFAT5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0072573 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of osteoclast proliferation(GO:0090291) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.2 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.1 | 0.2 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 | 0.2 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.0 | 0.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.1 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.0 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0021527 | skeletal muscle satellite cell differentiation(GO:0014816) spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.0 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.0 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.3 | GO:0030914 | SAGA complex(GO:0000124) STAGA complex(GO:0030914) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031896 | vasopressin receptor binding(GO:0031893) V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.2 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.0 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |