Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
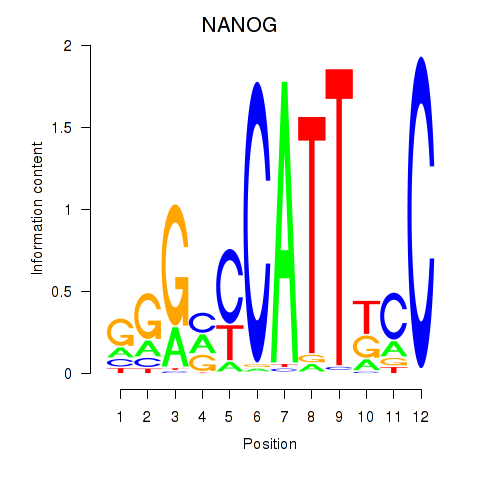
Results for NANOG
Z-value: 0.71
Transcription factors associated with NANOG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NANOG
|
ENSG00000111704.6 | Nanog homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NANOG | hg19_v2_chr12_+_7941989_7942014 | -0.09 | 8.6e-01 | Click! |
Activity profile of NANOG motif
Sorted Z-values of NANOG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_93692681 | 0.52 |
ENST00000348974.4
|
PROS1
|
protein S (alpha) |
| chrX_-_107019181 | 0.35 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr10_+_5566916 | 0.35 |
ENST00000315238.1
|
CALML3
|
calmodulin-like 3 |
| chr12_-_123215306 | 0.34 |
ENST00000356987.2
ENST00000436083.2 |
HCAR1
|
hydroxycarboxylic acid receptor 1 |
| chr19_+_6372444 | 0.34 |
ENST00000245812.3
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr3_-_93692781 | 0.31 |
ENST00000394236.3
|
PROS1
|
protein S (alpha) |
| chr6_+_26087509 | 0.28 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr2_-_112237835 | 0.27 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr8_-_27468717 | 0.26 |
ENST00000520796.1
ENST00000520491.1 |
CLU
|
clusterin |
| chr8_-_27469383 | 0.26 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr8_-_42751820 | 0.25 |
ENST00000526349.1
ENST00000527424.1 ENST00000534961.1 ENST00000319073.4 |
RNF170
|
ring finger protein 170 |
| chr2_+_87769459 | 0.25 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr11_+_1295809 | 0.25 |
ENST00000598274.1
|
AC136297.1
|
Uncharacterized protein |
| chr13_+_108921977 | 0.24 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr8_-_101733794 | 0.24 |
ENST00000523555.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr3_+_121774202 | 0.24 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr1_+_173837488 | 0.24 |
ENST00000427304.1
ENST00000432989.1 ENST00000367702.1 |
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr1_-_116383738 | 0.24 |
ENST00000320238.3
|
NHLH2
|
nescient helix loop helix 2 |
| chr6_-_33714667 | 0.22 |
ENST00000293756.4
|
IP6K3
|
inositol hexakisphosphate kinase 3 |
| chr4_-_140544386 | 0.21 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr17_-_56065540 | 0.20 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr2_+_149402009 | 0.20 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr3_+_23847432 | 0.20 |
ENST00000346855.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr4_+_154074217 | 0.20 |
ENST00000437508.2
|
TRIM2
|
tripartite motif containing 2 |
| chr13_+_100258907 | 0.20 |
ENST00000376355.3
ENST00000376360.1 ENST00000444838.2 ENST00000376354.1 ENST00000339105.4 |
CLYBL
|
citrate lyase beta like |
| chr6_+_26087646 | 0.20 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr10_-_128110441 | 0.19 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr7_+_97840739 | 0.19 |
ENST00000609256.1
|
BHLHA15
|
basic helix-loop-helix family, member a15 |
| chr12_-_46663734 | 0.18 |
ENST00000550173.1
|
SLC38A1
|
solute carrier family 38, member 1 |
| chr8_-_27469196 | 0.18 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr2_+_171673072 | 0.18 |
ENST00000358196.3
ENST00000375272.1 |
GAD1
|
glutamate decarboxylase 1 (brain, 67kDa) |
| chr10_+_64133934 | 0.17 |
ENST00000395254.3
ENST00000395255.3 ENST00000410046.3 |
ZNF365
|
zinc finger protein 365 |
| chr6_+_90272488 | 0.17 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr6_-_20212630 | 0.17 |
ENST00000324607.7
ENST00000541730.1 ENST00000536798.1 |
MBOAT1
|
membrane bound O-acyltransferase domain containing 1 |
| chr7_+_102715315 | 0.17 |
ENST00000428183.2
ENST00000323716.3 ENST00000441711.2 ENST00000454559.1 ENST00000425331.1 ENST00000541300.1 |
ARMC10
|
armadillo repeat containing 10 |
| chr6_-_33714752 | 0.17 |
ENST00000451316.1
|
IP6K3
|
inositol hexakisphosphate kinase 3 |
| chr3_-_149093499 | 0.16 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr12_-_42878101 | 0.16 |
ENST00000552108.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr16_+_68573116 | 0.16 |
ENST00000570495.1
ENST00000563169.2 ENST00000564323.1 ENST00000562156.1 ENST00000573685.1 |
ZFP90
|
ZFP90 zinc finger protein |
| chr7_-_99277610 | 0.16 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr10_-_70092671 | 0.16 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr8_-_27468945 | 0.15 |
ENST00000405140.3
|
CLU
|
clusterin |
| chr8_-_27468842 | 0.15 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr10_-_118885954 | 0.15 |
ENST00000392901.4
|
KIAA1598
|
KIAA1598 |
| chr20_+_43343517 | 0.14 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr6_+_148663729 | 0.14 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr3_+_32147997 | 0.14 |
ENST00000282541.5
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr1_+_210406121 | 0.14 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr16_+_68573640 | 0.14 |
ENST00000398253.2
ENST00000573161.1 |
ZFP90
|
ZFP90 zinc finger protein |
| chr8_-_101730061 | 0.13 |
ENST00000519100.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr12_+_19389814 | 0.13 |
ENST00000536974.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chrX_+_134124968 | 0.13 |
ENST00000330288.4
|
SMIM10
|
small integral membrane protein 10 |
| chr12_+_7456880 | 0.13 |
ENST00000399422.4
|
ACSM4
|
acyl-CoA synthetase medium-chain family member 4 |
| chr2_-_175113088 | 0.13 |
ENST00000409546.1
ENST00000428402.2 |
OLA1
|
Obg-like ATPase 1 |
| chr20_-_3762087 | 0.13 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr3_+_107244229 | 0.13 |
ENST00000456419.1
ENST00000402163.2 |
BBX
|
bobby sox homolog (Drosophila) |
| chr5_-_169816638 | 0.13 |
ENST00000521859.1
ENST00000274629.4 |
KCNMB1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr3_+_32148106 | 0.12 |
ENST00000425459.1
ENST00000431009.1 |
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr6_+_163837347 | 0.12 |
ENST00000544436.1
|
QKI
|
QKI, KH domain containing, RNA binding |
| chr6_+_155537771 | 0.12 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr4_+_154073469 | 0.12 |
ENST00000441616.1
|
TRIM2
|
tripartite motif containing 2 |
| chr3_+_132379154 | 0.11 |
ENST00000468022.1
ENST00000473651.1 ENST00000494238.2 |
UBA5
|
ubiquitin-like modifier activating enzyme 5 |
| chrX_+_17653413 | 0.11 |
ENST00000398097.3
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr2_+_170590321 | 0.11 |
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23 |
| chr6_-_119031228 | 0.11 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr10_+_123923205 | 0.11 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr20_+_43343476 | 0.11 |
ENST00000372868.2
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr20_+_43343886 | 0.11 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr3_-_15469045 | 0.11 |
ENST00000450816.2
|
METTL6
|
methyltransferase like 6 |
| chr12_+_69080734 | 0.11 |
ENST00000378905.2
|
NUP107
|
nucleoporin 107kDa |
| chr3_+_38388251 | 0.11 |
ENST00000427323.1
ENST00000207870.3 ENST00000542835.1 |
XYLB
|
xylulokinase homolog (H. influenzae) |
| chr6_+_107077471 | 0.11 |
ENST00000369044.1
|
QRSL1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr19_-_13213662 | 0.10 |
ENST00000264824.4
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr6_+_292051 | 0.10 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr17_+_7905912 | 0.10 |
ENST00000254854.4
|
GUCY2D
|
guanylate cyclase 2D, membrane (retina-specific) |
| chr1_+_47264711 | 0.10 |
ENST00000371923.4
ENST00000271153.4 ENST00000371919.4 |
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1 |
| chr15_-_50978965 | 0.10 |
ENST00000560955.1
ENST00000313478.7 |
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr2_-_97405775 | 0.10 |
ENST00000264963.4
ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L
|
lectin, mannose-binding 2-like |
| chr12_-_69080590 | 0.10 |
ENST00000433116.2
ENST00000500695.2 |
RP11-637A17.2
|
RP11-637A17.2 |
| chr10_-_70092635 | 0.10 |
ENST00000309049.4
|
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr15_+_78833105 | 0.09 |
ENST00000558341.1
ENST00000559437.1 |
PSMA4
|
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr6_+_31540056 | 0.09 |
ENST00000418386.2
|
LTA
|
lymphotoxin alpha |
| chr1_-_145589424 | 0.09 |
ENST00000334513.5
|
NUDT17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr10_+_123923105 | 0.09 |
ENST00000368999.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr3_-_120461378 | 0.09 |
ENST00000273375.3
|
RABL3
|
RAB, member of RAS oncogene family-like 3 |
| chr14_-_98444369 | 0.09 |
ENST00000554822.1
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr6_+_107077435 | 0.09 |
ENST00000369046.4
|
QRSL1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr19_+_37825614 | 0.09 |
ENST00000591259.1
ENST00000590582.1 |
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr7_+_131012605 | 0.09 |
ENST00000446815.1
ENST00000352689.6 |
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr16_-_30997533 | 0.09 |
ENST00000602217.1
|
AC135048.1
|
Uncharacterized protein |
| chr16_+_3508096 | 0.09 |
ENST00000577013.1
ENST00000570819.1 |
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr6_+_88032299 | 0.08 |
ENST00000608353.1
ENST00000392863.1 ENST00000229570.5 ENST00000608525.1 ENST00000608868.1 |
SMIM8
|
small integral membrane protein 8 |
| chrX_-_118827333 | 0.08 |
ENST00000360156.7
ENST00000354228.4 ENST00000489216.1 ENST00000354416.3 ENST00000394610.1 ENST00000343984.5 |
SEPT6
|
septin 6 |
| chrX_+_103173457 | 0.08 |
ENST00000419165.1
|
TMSB15B
|
thymosin beta 15B |
| chr6_+_52226897 | 0.08 |
ENST00000442253.2
|
PAQR8
|
progestin and adipoQ receptor family member VIII |
| chr22_+_37447771 | 0.08 |
ENST00000402077.3
ENST00000403888.3 ENST00000456470.1 |
KCTD17
|
potassium channel tetramerization domain containing 17 |
| chr3_-_155572164 | 0.08 |
ENST00000392845.3
ENST00000359479.3 |
SLC33A1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr15_+_44580955 | 0.08 |
ENST00000345795.2
ENST00000360824.3 |
CASC4
|
cancer susceptibility candidate 4 |
| chr2_-_240322685 | 0.08 |
ENST00000544989.1
|
HDAC4
|
histone deacetylase 4 |
| chr15_+_78832747 | 0.08 |
ENST00000560217.1
ENST00000044462.7 ENST00000559082.1 ENST00000559948.1 ENST00000413382.2 ENST00000559146.1 ENST00000558281.1 |
PSMA4
|
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr11_-_14379997 | 0.08 |
ENST00000526063.1
ENST00000532814.1 |
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chrX_-_16888276 | 0.08 |
ENST00000493145.1
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr2_-_214016314 | 0.08 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr16_-_70719925 | 0.08 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr1_+_173837214 | 0.07 |
ENST00000367704.1
|
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr10_+_123922941 | 0.07 |
ENST00000360561.3
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr6_+_116850174 | 0.07 |
ENST00000416171.2
ENST00000368597.2 ENST00000452373.1 ENST00000405399.1 |
FAM26D
|
family with sequence similarity 26, member D |
| chr3_-_120461353 | 0.07 |
ENST00000483733.1
|
RABL3
|
RAB, member of RAS oncogene family-like 3 |
| chr3_+_15469058 | 0.07 |
ENST00000432764.2
|
EAF1
|
ELL associated factor 1 |
| chr16_+_14280742 | 0.07 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr19_+_42381173 | 0.07 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr10_-_27443155 | 0.07 |
ENST00000427324.1
ENST00000326799.3 |
YME1L1
|
YME1-like 1 ATPase |
| chr14_+_101295948 | 0.07 |
ENST00000452514.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr7_-_76829125 | 0.07 |
ENST00000248598.5
|
FGL2
|
fibrinogen-like 2 |
| chrX_-_19765692 | 0.07 |
ENST00000432234.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr2_-_175113301 | 0.06 |
ENST00000344357.5
ENST00000284719.3 |
OLA1
|
Obg-like ATPase 1 |
| chr1_+_35734562 | 0.06 |
ENST00000314607.6
ENST00000373297.2 |
ZMYM4
|
zinc finger, MYM-type 4 |
| chr15_-_78526942 | 0.06 |
ENST00000258873.4
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr18_-_23671139 | 0.06 |
ENST00000579061.1
ENST00000542420.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr5_+_138089100 | 0.06 |
ENST00000520339.1
ENST00000355078.5 ENST00000302763.7 ENST00000518910.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr6_-_107077347 | 0.06 |
ENST00000369063.3
ENST00000539449.1 |
RTN4IP1
|
reticulon 4 interacting protein 1 |
| chr7_-_28220354 | 0.06 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr3_-_53925863 | 0.06 |
ENST00000541726.1
ENST00000495461.1 |
SELK
|
Selenoprotein K |
| chr17_-_43128943 | 0.06 |
ENST00000588499.1
ENST00000593094.1 |
DCAKD
|
dephospho-CoA kinase domain containing |
| chr4_-_108204846 | 0.06 |
ENST00000513208.1
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr1_+_161551101 | 0.06 |
ENST00000367962.4
ENST00000367960.5 ENST00000403078.3 ENST00000428605.2 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr3_-_15469006 | 0.06 |
ENST00000443029.1
ENST00000383790.3 ENST00000383789.5 |
METTL6
|
methyltransferase like 6 |
| chr6_-_30524951 | 0.05 |
ENST00000376621.3
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr19_-_36870087 | 0.05 |
ENST00000270001.7
|
ZFP14
|
ZFP14 zinc finger protein |
| chr1_-_72748140 | 0.05 |
ENST00000434200.1
|
NEGR1
|
neuronal growth regulator 1 |
| chr4_-_5891918 | 0.05 |
ENST00000512574.1
|
CRMP1
|
collapsin response mediator protein 1 |
| chr12_-_31743901 | 0.05 |
ENST00000354285.4
|
DENND5B
|
DENN/MADD domain containing 5B |
| chr14_-_51297837 | 0.05 |
ENST00000245441.5
ENST00000389868.3 ENST00000382041.3 ENST00000324330.9 ENST00000453196.1 ENST00000453401.2 |
NIN
|
ninein (GSK3B interacting protein) |
| chr9_+_124030338 | 0.05 |
ENST00000449773.1
ENST00000432226.1 ENST00000436847.1 ENST00000394353.2 ENST00000449733.1 ENST00000412819.1 ENST00000341272.2 ENST00000373808.2 |
GSN
|
gelsolin |
| chr10_+_69644404 | 0.05 |
ENST00000212015.6
|
SIRT1
|
sirtuin 1 |
| chr6_+_90272027 | 0.04 |
ENST00000522441.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr17_+_43224684 | 0.04 |
ENST00000332499.2
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1 |
| chr18_+_3262954 | 0.04 |
ENST00000584539.1
|
MYL12B
|
myosin, light chain 12B, regulatory |
| chr6_+_32407619 | 0.04 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr5_+_156693159 | 0.04 |
ENST00000347377.6
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr1_+_104197912 | 0.04 |
ENST00000430659.1
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr12_-_25055177 | 0.04 |
ENST00000538118.1
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chrX_-_133931164 | 0.04 |
ENST00000370790.1
ENST00000298090.6 |
FAM122B
|
family with sequence similarity 122B |
| chr14_-_92198403 | 0.04 |
ENST00000553329.1
ENST00000256343.3 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr14_+_23791159 | 0.04 |
ENST00000557702.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr15_+_77713222 | 0.04 |
ENST00000558176.1
|
HMG20A
|
high mobility group 20A |
| chr3_+_44916098 | 0.04 |
ENST00000296125.4
|
TGM4
|
transglutaminase 4 |
| chr15_+_44580899 | 0.04 |
ENST00000559222.1
ENST00000299957.6 |
CASC4
|
cancer susceptibility candidate 4 |
| chr6_-_136610911 | 0.04 |
ENST00000530767.1
ENST00000527759.1 ENST00000527536.1 ENST00000529826.1 ENST00000531224.1 ENST00000353331.4 |
BCLAF1
|
BCL2-associated transcription factor 1 |
| chr5_-_55290773 | 0.03 |
ENST00000502326.3
ENST00000381298.2 |
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chr16_+_3508063 | 0.03 |
ENST00000576787.1
ENST00000572942.1 ENST00000576916.1 ENST00000575076.1 ENST00000572131.1 |
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr15_+_90319557 | 0.03 |
ENST00000341735.3
|
MESP2
|
mesoderm posterior 2 homolog (mouse) |
| chr1_+_203096831 | 0.03 |
ENST00000337894.4
|
ADORA1
|
adenosine A1 receptor |
| chr2_-_97536490 | 0.03 |
ENST00000449330.1
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr22_+_42095497 | 0.03 |
ENST00000401548.3
ENST00000540833.1 ENST00000400107.1 ENST00000300398.4 |
MEI1
|
meiosis inhibitor 1 |
| chr2_+_45878407 | 0.03 |
ENST00000421201.1
|
PRKCE
|
protein kinase C, epsilon |
| chr10_-_1034237 | 0.03 |
ENST00000381466.1
|
AL359878.1
|
Uncharacterized protein |
| chr1_+_44870866 | 0.03 |
ENST00000355387.2
ENST00000361799.2 |
RNF220
|
ring finger protein 220 |
| chr10_+_129845823 | 0.02 |
ENST00000306042.5
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr10_-_79686284 | 0.02 |
ENST00000372391.2
ENST00000372388.2 |
DLG5
|
discs, large homolog 5 (Drosophila) |
| chr8_+_9413410 | 0.02 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr14_-_98444438 | 0.02 |
ENST00000512901.2
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr19_-_39926555 | 0.02 |
ENST00000599539.1
ENST00000339471.4 ENST00000601655.1 ENST00000251453.3 |
RPS16
|
ribosomal protein S16 |
| chr10_+_72575643 | 0.02 |
ENST00000373202.3
|
SGPL1
|
sphingosine-1-phosphate lyase 1 |
| chr17_+_75372165 | 0.02 |
ENST00000427674.2
|
SEPT9
|
septin 9 |
| chr3_+_15468862 | 0.02 |
ENST00000396842.2
|
EAF1
|
ELL associated factor 1 |
| chrX_-_118827113 | 0.02 |
ENST00000394617.2
|
SEPT6
|
septin 6 |
| chr7_+_134576317 | 0.02 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr5_+_32585605 | 0.02 |
ENST00000265073.4
ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr2_+_219081817 | 0.02 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr17_-_56065484 | 0.02 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr16_+_31494323 | 0.02 |
ENST00000569576.1
ENST00000330498.3 |
SLC5A2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr1_+_203097407 | 0.02 |
ENST00000367235.1
|
ADORA1
|
adenosine A1 receptor |
| chr9_-_99381660 | 0.02 |
ENST00000375240.3
ENST00000463569.1 |
CDC14B
|
cell division cycle 14B |
| chr12_-_85306562 | 0.01 |
ENST00000551612.1
ENST00000450363.3 ENST00000552192.1 |
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr2_-_69664586 | 0.01 |
ENST00000303698.3
ENST00000394305.1 ENST00000410022.2 |
NFU1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr4_-_16077741 | 0.01 |
ENST00000447510.2
ENST00000540805.1 ENST00000539194.1 |
PROM1
|
prominin 1 |
| chr7_-_102715263 | 0.01 |
ENST00000379305.3
|
FBXL13
|
F-box and leucine-rich repeat protein 13 |
| chrX_+_47092314 | 0.01 |
ENST00000218348.3
|
USP11
|
ubiquitin specific peptidase 11 |
| chr3_-_134369853 | 0.01 |
ENST00000508956.1
ENST00000503669.1 ENST00000423778.2 |
KY
|
kyphoscoliosis peptidase |
| chr7_-_143991230 | 0.01 |
ENST00000543357.1
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35 |
| chr17_-_55911970 | 0.01 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr21_-_34960930 | 0.01 |
ENST00000437395.1
|
DONSON
|
downstream neighbor of SON |
| chr6_-_43021612 | 0.01 |
ENST00000535468.1
|
CUL7
|
cullin 7 |
| chr9_+_71819927 | 0.01 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr8_+_142264664 | 0.01 |
ENST00000518520.1
|
RP11-10J21.3
|
Uncharacterized protein |
| chr18_+_3262415 | 0.01 |
ENST00000581193.1
ENST00000400175.5 |
MYL12B
|
myosin, light chain 12B, regulatory |
| chr19_-_8070474 | 0.01 |
ENST00000407627.2
ENST00000593807.1 |
ELAVL1
|
ELAV like RNA binding protein 1 |
| chr5_+_156693091 | 0.01 |
ENST00000318218.6
ENST00000442283.2 ENST00000522463.1 ENST00000521420.1 |
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr4_+_3344141 | 0.01 |
ENST00000306648.7
|
RGS12
|
regulator of G-protein signaling 12 |
| chr5_+_68513557 | 0.01 |
ENST00000256441.4
|
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr20_+_37554955 | 0.00 |
ENST00000217429.4
|
FAM83D
|
family with sequence similarity 83, member D |
| chr5_-_33892204 | 0.00 |
ENST00000504830.1
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr12_-_52946923 | 0.00 |
ENST00000267119.5
|
KRT71
|
keratin 71 |
| chr10_-_69455873 | 0.00 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr7_+_150382781 | 0.00 |
ENST00000223293.5
ENST00000474605.1 |
GIMAP2
|
GTPase, IMAP family member 2 |
| chr6_-_31633402 | 0.00 |
ENST00000375893.2
|
GPANK1
|
G patch domain and ankyrin repeats 1 |
| chr1_+_161632937 | 0.00 |
ENST00000236937.9
ENST00000367961.4 ENST00000358671.5 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr15_+_78833071 | 0.00 |
ENST00000559365.1
|
PSMA4
|
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr4_-_16900184 | 0.00 |
ENST00000515064.1
|
LDB2
|
LIM domain binding 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NANOG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.1 | 1.0 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.3 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.2 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.2 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.1 | 0.3 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 0.8 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 | 0.2 | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.1 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.0 | GO:0042323 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.1 | GO:0002605 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.0 | 0.1 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.0 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.0 | 0.0 | GO:1990619 | histone H3-K9 deacetylation(GO:1990619) |
| 0.0 | 0.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.0 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.5 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0005940 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.4 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 1.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |