Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
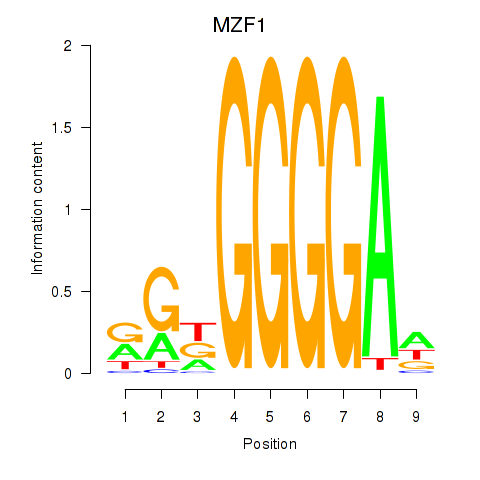
Results for MZF1
Z-value: 0.43
Transcription factors associated with MZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MZF1
|
ENSG00000099326.4 | myeloid zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MZF1 | hg19_v2_chr19_-_59084647_59084721 | 0.75 | 8.4e-02 | Click! |
Activity profile of MZF1 motif
Sorted Z-values of MZF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_61765315 | 0.36 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr3_-_169381166 | 0.35 |
ENST00000486748.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr19_-_10628098 | 0.30 |
ENST00000590601.1
|
S1PR5
|
sphingosine-1-phosphate receptor 5 |
| chr4_-_56412713 | 0.30 |
ENST00000435527.2
|
CLOCK
|
clock circadian regulator |
| chr2_+_206950095 | 0.25 |
ENST00000435627.1
|
AC007383.3
|
AC007383.3 |
| chr14_-_71107921 | 0.25 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chrX_-_108868390 | 0.24 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr5_+_115177178 | 0.22 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr3_+_119814070 | 0.21 |
ENST00000469070.1
|
RP11-18H7.1
|
RP11-18H7.1 |
| chr17_+_2264983 | 0.20 |
ENST00000574650.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr18_-_53255766 | 0.19 |
ENST00000566286.1
ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4
|
transcription factor 4 |
| chr1_+_6845578 | 0.19 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chrX_-_119694538 | 0.19 |
ENST00000371322.5
|
CUL4B
|
cullin 4B |
| chr14_-_36990061 | 0.18 |
ENST00000546983.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr17_-_56065540 | 0.18 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr5_-_81046904 | 0.18 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr12_+_65996599 | 0.17 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr8_+_145490549 | 0.17 |
ENST00000340695.2
|
SCXA
|
scleraxis homolog A (mouse) |
| chr1_-_167906277 | 0.17 |
ENST00000271373.4
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr20_+_2083540 | 0.17 |
ENST00000400064.3
|
STK35
|
serine/threonine kinase 35 |
| chr1_-_11115877 | 0.16 |
ENST00000490101.1
|
SRM
|
spermidine synthase |
| chr17_-_36904437 | 0.16 |
ENST00000585100.1
ENST00000360797.2 ENST00000578109.1 ENST00000579882.1 |
PCGF2
|
polycomb group ring finger 2 |
| chr17_+_42634844 | 0.15 |
ENST00000315323.3
|
FZD2
|
frizzled family receptor 2 |
| chr3_-_93692681 | 0.14 |
ENST00000348974.4
|
PROS1
|
protein S (alpha) |
| chr15_-_83224682 | 0.14 |
ENST00000562833.1
|
RP11-152F13.10
|
RP11-152F13.10 |
| chr7_-_112579869 | 0.14 |
ENST00000297145.4
|
C7orf60
|
chromosome 7 open reading frame 60 |
| chr2_+_145780767 | 0.14 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr13_-_52026730 | 0.14 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr16_-_79634595 | 0.14 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr3_-_12200851 | 0.14 |
ENST00000287814.4
|
TIMP4
|
TIMP metallopeptidase inhibitor 4 |
| chr1_+_78470530 | 0.14 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr5_-_81046841 | 0.14 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr15_+_75640068 | 0.13 |
ENST00000565051.1
ENST00000564257.1 ENST00000567005.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr14_-_76447494 | 0.13 |
ENST00000238682.3
|
TGFB3
|
transforming growth factor, beta 3 |
| chr3_-_88108192 | 0.13 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr1_+_167599532 | 0.13 |
ENST00000537350.1
|
RCSD1
|
RCSD domain containing 1 |
| chr15_-_71055769 | 0.13 |
ENST00000539319.1
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr7_-_79082867 | 0.13 |
ENST00000419488.1
ENST00000354212.4 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr11_-_94965667 | 0.12 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr2_-_26251481 | 0.12 |
ENST00000599234.1
|
AC013449.1
|
Uncharacterized protein |
| chr7_-_127032363 | 0.12 |
ENST00000393312.1
|
ZNF800
|
zinc finger protein 800 |
| chr6_-_31869769 | 0.12 |
ENST00000375527.2
|
ZBTB12
|
zinc finger and BTB domain containing 12 |
| chr5_-_81046922 | 0.12 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr4_+_30721968 | 0.12 |
ENST00000361762.2
|
PCDH7
|
protocadherin 7 |
| chr2_+_149402989 | 0.12 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr9_+_34653861 | 0.12 |
ENST00000556792.1
ENST00000318041.9 ENST00000378817.4 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr14_-_77495007 | 0.12 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr17_+_38465441 | 0.12 |
ENST00000577646.1
ENST00000254066.5 |
RARA
|
retinoic acid receptor, alpha |
| chr17_+_29421987 | 0.12 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr17_-_49337392 | 0.12 |
ENST00000376381.2
ENST00000586178.1 |
MBTD1
|
mbt domain containing 1 |
| chr11_+_86748957 | 0.12 |
ENST00000526733.1
ENST00000532959.1 |
TMEM135
|
transmembrane protein 135 |
| chr5_-_149465990 | 0.12 |
ENST00000543093.1
|
CSF1R
|
colony stimulating factor 1 receptor |
| chr1_-_67390474 | 0.12 |
ENST00000371023.3
ENST00000371022.3 ENST00000371026.3 ENST00000431318.1 |
WDR78
|
WD repeat domain 78 |
| chr12_+_57522439 | 0.12 |
ENST00000338962.4
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr17_+_26369865 | 0.12 |
ENST00000582037.1
|
NLK
|
nemo-like kinase |
| chr6_-_86353510 | 0.12 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr12_+_57522692 | 0.11 |
ENST00000554174.1
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr10_+_102672712 | 0.11 |
ENST00000370271.3
ENST00000370269.3 ENST00000609386.1 |
FAM178A
|
family with sequence similarity 178, member A |
| chr19_+_13135439 | 0.11 |
ENST00000586873.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr2_-_61765732 | 0.11 |
ENST00000443240.1
ENST00000436018.1 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr4_+_184427235 | 0.11 |
ENST00000412117.1
ENST00000434682.2 |
ING2
|
inhibitor of growth family, member 2 |
| chr18_-_658244 | 0.11 |
ENST00000585033.1
ENST00000323813.3 |
C18orf56
|
chromosome 18 open reading frame 56 |
| chr22_-_31742218 | 0.11 |
ENST00000266269.5
ENST00000405309.3 ENST00000351933.4 |
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr22_+_40440804 | 0.11 |
ENST00000441751.1
ENST00000301923.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr11_+_130184888 | 0.11 |
ENST00000602376.1
ENST00000532116.3 ENST00000602310.1 |
RP11-121M22.1
|
RP11-121M22.1 |
| chr5_+_32531893 | 0.10 |
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr8_+_22250334 | 0.10 |
ENST00000520832.1
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr7_-_112579673 | 0.10 |
ENST00000432572.1
|
C7orf60
|
chromosome 7 open reading frame 60 |
| chr11_+_86511549 | 0.10 |
ENST00000533902.2
|
PRSS23
|
protease, serine, 23 |
| chr15_-_74043816 | 0.10 |
ENST00000379822.4
|
C15orf59
|
chromosome 15 open reading frame 59 |
| chr2_+_46769798 | 0.10 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr10_+_21823243 | 0.10 |
ENST00000307729.7
ENST00000377091.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr14_+_32546145 | 0.10 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr7_+_114562172 | 0.10 |
ENST00000393486.1
ENST00000257724.3 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr11_-_35547277 | 0.10 |
ENST00000527605.1
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr7_-_127032741 | 0.10 |
ENST00000393313.1
ENST00000265827.3 ENST00000434602.1 |
ZNF800
|
zinc finger protein 800 |
| chr12_+_6881678 | 0.10 |
ENST00000441671.2
ENST00000203629.2 |
LAG3
|
lymphocyte-activation gene 3 |
| chrX_-_13835461 | 0.10 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr4_-_153274078 | 0.10 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr1_+_150229554 | 0.10 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr9_-_6007787 | 0.10 |
ENST00000399933.3
ENST00000381461.2 ENST00000513355.2 |
KIAA2026
|
KIAA2026 |
| chr1_-_159915386 | 0.10 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr1_+_6845497 | 0.10 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chrX_+_123095546 | 0.10 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr2_+_220325977 | 0.10 |
ENST00000396686.1
ENST00000396689.2 |
SPEG
|
SPEG complex locus |
| chr2_-_172017393 | 0.09 |
ENST00000442919.2
|
TLK1
|
tousled-like kinase 1 |
| chr3_-_168864315 | 0.09 |
ENST00000475754.1
ENST00000484519.1 |
MECOM
|
MDS1 and EVI1 complex locus |
| chr19_-_59084647 | 0.09 |
ENST00000594234.1
ENST00000596039.1 |
MZF1
|
myeloid zinc finger 1 |
| chr1_-_221915418 | 0.09 |
ENST00000323825.3
ENST00000366899.3 |
DUSP10
|
dual specificity phosphatase 10 |
| chr19_-_49371711 | 0.09 |
ENST00000355496.5
ENST00000263265.6 |
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr8_-_75233563 | 0.09 |
ENST00000342232.4
|
JPH1
|
junctophilin 1 |
| chr11_-_35547151 | 0.09 |
ENST00000378878.3
ENST00000529303.1 ENST00000278360.3 |
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr17_-_48943706 | 0.09 |
ENST00000499247.2
|
TOB1
|
transducer of ERBB2, 1 |
| chrX_-_119695279 | 0.09 |
ENST00000336592.6
|
CUL4B
|
cullin 4B |
| chr2_-_61697862 | 0.09 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr2_-_45236540 | 0.09 |
ENST00000303077.6
|
SIX2
|
SIX homeobox 2 |
| chr2_+_220325441 | 0.09 |
ENST00000396688.1
|
SPEG
|
SPEG complex locus |
| chr1_+_67390578 | 0.09 |
ENST00000371018.3
ENST00000355977.6 ENST00000357692.2 ENST00000401041.1 ENST00000371016.1 ENST00000371014.1 ENST00000371012.2 |
MIER1
|
mesoderm induction early response 1, transcriptional regulator |
| chr1_-_150208412 | 0.09 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr17_+_73089382 | 0.09 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr8_-_101733794 | 0.09 |
ENST00000523555.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chrX_+_123095860 | 0.09 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr11_+_86748863 | 0.09 |
ENST00000340353.7
|
TMEM135
|
transmembrane protein 135 |
| chr3_-_50360165 | 0.09 |
ENST00000428028.1
|
HYAL2
|
hyaluronoglucosaminidase 2 |
| chr12_-_7077125 | 0.09 |
ENST00000545555.2
|
PHB2
|
prohibitin 2 |
| chr3_+_110790590 | 0.08 |
ENST00000485303.1
|
PVRL3
|
poliovirus receptor-related 3 |
| chrX_-_135849484 | 0.08 |
ENST00000370620.1
ENST00000535227.1 |
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr10_+_21823079 | 0.08 |
ENST00000377100.3
ENST00000377072.3 ENST00000446906.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr14_-_64010046 | 0.08 |
ENST00000337537.3
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform |
| chr17_+_79990058 | 0.08 |
ENST00000584341.1
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr4_+_129730947 | 0.08 |
ENST00000452328.2
ENST00000504089.1 |
PHF17
|
jade family PHD finger 1 |
| chr16_+_22019404 | 0.08 |
ENST00000542527.2
ENST00000569656.1 ENST00000562695.1 |
C16orf52
|
chromosome 16 open reading frame 52 |
| chr17_+_29421900 | 0.08 |
ENST00000358273.4
ENST00000356175.3 |
NF1
|
neurofibromin 1 |
| chr11_+_86749035 | 0.08 |
ENST00000305494.5
ENST00000535167.1 |
TMEM135
|
transmembrane protein 135 |
| chr17_-_46667594 | 0.08 |
ENST00000476342.1
ENST00000460160.1 ENST00000472863.1 |
HOXB3
|
homeobox B3 |
| chr17_-_7193711 | 0.08 |
ENST00000571464.1
|
YBX2
|
Y box binding protein 2 |
| chr2_+_61108650 | 0.08 |
ENST00000295025.8
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr8_+_104831472 | 0.08 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr5_+_82767284 | 0.08 |
ENST00000265077.3
|
VCAN
|
versican |
| chr11_+_86511569 | 0.08 |
ENST00000441050.1
|
PRSS23
|
protease, serine, 23 |
| chr1_+_156052354 | 0.08 |
ENST00000368301.2
|
LMNA
|
lamin A/C |
| chr3_-_182698381 | 0.08 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chrX_+_123095890 | 0.08 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr2_+_46926326 | 0.08 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr11_+_47279504 | 0.08 |
ENST00000441012.2
ENST00000437276.1 ENST00000436029.1 ENST00000467728.1 ENST00000405853.3 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr1_+_160370344 | 0.08 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr19_+_13135790 | 0.08 |
ENST00000358552.3
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr14_+_32546485 | 0.08 |
ENST00000345122.3
ENST00000432921.1 ENST00000433497.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr17_+_39968926 | 0.08 |
ENST00000585664.1
ENST00000585922.1 ENST00000429461.1 |
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr13_+_76123883 | 0.08 |
ENST00000377595.3
|
UCHL3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr4_-_168155577 | 0.07 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr3_-_93692781 | 0.07 |
ENST00000394236.3
|
PROS1
|
protein S (alpha) |
| chr19_-_38916839 | 0.07 |
ENST00000433821.2
ENST00000426920.2 ENST00000587753.1 ENST00000454404.2 ENST00000293062.9 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr22_+_38321840 | 0.07 |
ENST00000454685.1
|
MICALL1
|
MICAL-like 1 |
| chr3_-_178865747 | 0.07 |
ENST00000435560.1
|
RP11-360P21.2
|
RP11-360P21.2 |
| chr16_-_67260901 | 0.07 |
ENST00000341546.3
ENST00000409509.1 ENST00000433915.1 ENST00000454102.2 |
LRRC29
AC040160.1
|
leucine rich repeat containing 29 Uncharacterized protein; cDNA FLJ57407, weakly similar to Mus musculus leucine rich repeat containing 29 (Lrrc29), mRNA |
| chr3_+_73045936 | 0.07 |
ENST00000356692.5
ENST00000488810.1 ENST00000394284.3 ENST00000295862.9 ENST00000495566.1 |
PPP4R2
|
protein phosphatase 4, regulatory subunit 2 |
| chr20_-_42816206 | 0.07 |
ENST00000372980.3
|
JPH2
|
junctophilin 2 |
| chr15_-_71055878 | 0.07 |
ENST00000322954.6
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr8_-_38126635 | 0.07 |
ENST00000529359.1
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr19_-_42759300 | 0.07 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr9_-_122131696 | 0.07 |
ENST00000373964.2
ENST00000265922.3 |
BRINP1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr19_+_34745442 | 0.07 |
ENST00000299505.6
ENST00000588470.1 ENST00000589583.1 ENST00000588338.2 |
KIAA0355
|
KIAA0355 |
| chr6_-_52860171 | 0.07 |
ENST00000370963.4
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr1_-_113162040 | 0.07 |
ENST00000358039.4
ENST00000369668.2 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr6_+_87865262 | 0.07 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr8_-_21988558 | 0.07 |
ENST00000312841.8
|
HR
|
hair growth associated |
| chr14_+_32546274 | 0.07 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr3_-_169381183 | 0.07 |
ENST00000494292.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr16_+_28505955 | 0.07 |
ENST00000564831.1
ENST00000328423.5 ENST00000431282.1 |
APOBR
|
apolipoprotein B receptor |
| chr2_+_219283815 | 0.07 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr4_+_140222609 | 0.07 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr18_+_19321281 | 0.07 |
ENST00000261537.6
|
MIB1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr3_-_48601206 | 0.07 |
ENST00000273610.3
|
UCN2
|
urocortin 2 |
| chr10_+_98592009 | 0.07 |
ENST00000540664.1
ENST00000371103.3 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chr4_+_174089904 | 0.07 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr2_+_121493717 | 0.07 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr3_+_110790715 | 0.07 |
ENST00000319792.3
|
PVRL3
|
poliovirus receptor-related 3 |
| chr9_-_74525847 | 0.07 |
ENST00000377041.2
|
ABHD17B
|
abhydrolase domain containing 17B |
| chr9_-_119449483 | 0.07 |
ENST00000288520.5
ENST00000358637.4 ENST00000341734.4 |
ASTN2
|
astrotactin 2 |
| chr7_+_130794878 | 0.07 |
ENST00000416992.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chrX_-_109561294 | 0.07 |
ENST00000372059.2
ENST00000262844.5 |
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr1_+_93913665 | 0.07 |
ENST00000271234.7
ENST00000370256.4 ENST00000260506.8 |
FNBP1L
|
formin binding protein 1-like |
| chr15_-_33447055 | 0.07 |
ENST00000559047.1
ENST00000561249.1 |
FMN1
|
formin 1 |
| chr11_-_96076334 | 0.07 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr15_-_49103184 | 0.06 |
ENST00000399334.3
ENST00000325747.5 |
CEP152
|
centrosomal protein 152kDa |
| chr1_+_154301264 | 0.06 |
ENST00000341822.2
|
ATP8B2
|
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr5_+_49962495 | 0.06 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr17_+_38278530 | 0.06 |
ENST00000398532.4
|
MSL1
|
male-specific lethal 1 homolog (Drosophila) |
| chr2_-_206950781 | 0.06 |
ENST00000403263.1
|
INO80D
|
INO80 complex subunit D |
| chr13_+_95364963 | 0.06 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chr14_-_36990354 | 0.06 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr15_-_37392724 | 0.06 |
ENST00000424352.2
|
MEIS2
|
Meis homeobox 2 |
| chr19_-_4065730 | 0.06 |
ENST00000601588.1
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr15_-_66084621 | 0.06 |
ENST00000564674.1
|
DENND4A
|
DENN/MADD domain containing 4A |
| chr13_-_31736027 | 0.06 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr5_-_94620239 | 0.06 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr1_+_24117662 | 0.06 |
ENST00000420982.1
ENST00000374505.2 |
LYPLA2
|
lysophospholipase II |
| chr2_+_45878407 | 0.06 |
ENST00000421201.1
|
PRKCE
|
protein kinase C, epsilon |
| chr3_+_9944303 | 0.06 |
ENST00000421412.1
ENST00000295980.3 |
IL17RE
|
interleukin 17 receptor E |
| chr16_-_67260691 | 0.06 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr1_-_53018654 | 0.06 |
ENST00000257177.4
ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr17_-_37764128 | 0.06 |
ENST00000302584.4
|
NEUROD2
|
neuronal differentiation 2 |
| chr4_-_84035905 | 0.06 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr4_-_153457197 | 0.06 |
ENST00000281708.4
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr7_-_100061869 | 0.06 |
ENST00000332375.3
|
C7orf61
|
chromosome 7 open reading frame 61 |
| chr19_-_46272462 | 0.06 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr19_+_3708338 | 0.06 |
ENST00000590545.1
|
TJP3
|
tight junction protein 3 |
| chr4_+_88928777 | 0.06 |
ENST00000237596.2
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant) |
| chr4_-_76598296 | 0.06 |
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr8_+_95653840 | 0.06 |
ENST00000520385.1
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr17_+_46132037 | 0.06 |
ENST00000582155.1
ENST00000583378.1 ENST00000536222.1 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr12_+_53443963 | 0.06 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr22_-_46373004 | 0.06 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chr8_+_55466915 | 0.06 |
ENST00000522711.2
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr1_+_89149905 | 0.06 |
ENST00000316005.7
ENST00000370521.3 ENST00000370505.3 |
PKN2
|
protein kinase N2 |
| chr7_-_17980091 | 0.06 |
ENST00000409389.1
ENST00000409604.1 ENST00000428135.3 |
SNX13
|
sorting nexin 13 |
| chr3_+_181429704 | 0.06 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr15_-_51058005 | 0.06 |
ENST00000261854.5
|
SPPL2A
|
signal peptide peptidase like 2A |
| chr4_-_89205879 | 0.06 |
ENST00000608933.1
ENST00000315194.4 ENST00000514204.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
Network of associatons between targets according to the STRING database.
First level regulatory network of MZF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.2 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.1 | GO:0042704 | detection of oxygen(GO:0003032) uterine wall breakdown(GO:0042704) |
| 0.0 | 0.3 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.0 | 0.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.2 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.1 | GO:1904799 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.1 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) regulation of respiratory burst involved in inflammatory response(GO:0060264) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.0 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.0 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.2 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.1 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) transformation of host cell by virus(GO:0019087) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.0 | GO:1904247 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.3 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.0 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.0 | GO:0002588 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.0 | 0.0 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.0 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.0 | GO:0072179 | regulation of pronephros size(GO:0035565) pronephric nephron tubule development(GO:0039020) nephric duct formation(GO:0072179) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.0 | 0.0 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.1 | 0.2 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.0 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.1 | GO:0048763 | HLH domain binding(GO:0043398) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.3 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.0 | GO:0031433 | telethonin binding(GO:0031433) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |