Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
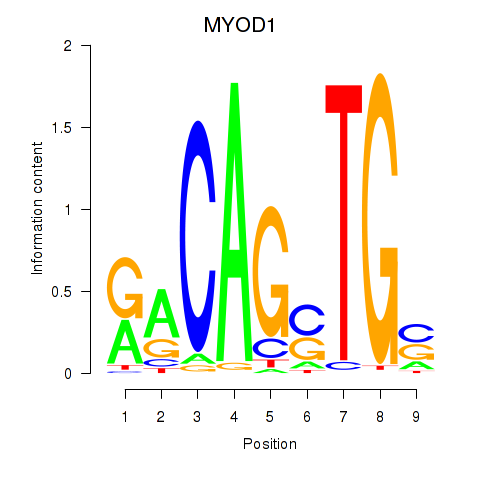
Results for MYOD1
Z-value: 0.67
Transcription factors associated with MYOD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYOD1
|
ENSG00000129152.3 | myogenic differentiation 1 |
Activity profile of MYOD1 motif
Sorted Z-values of MYOD1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_39258461 | 0.63 |
ENST00000440582.1
|
KRTAP4-16P
|
keratin associated protein 4-16, pseudogene |
| chr1_-_47655686 | 0.48 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr12_-_58145889 | 0.40 |
ENST00000547853.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr14_-_100625932 | 0.36 |
ENST00000553834.1
|
DEGS2
|
delta(4)-desaturase, sphingolipid 2 |
| chr3_+_32148106 | 0.34 |
ENST00000425459.1
ENST00000431009.1 |
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr12_+_56473628 | 0.28 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr7_+_66800928 | 0.26 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr17_+_38119216 | 0.26 |
ENST00000301659.4
|
GSDMA
|
gasdermin A |
| chrX_+_70521584 | 0.26 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr1_-_201346761 | 0.26 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr5_+_131630117 | 0.25 |
ENST00000200652.3
|
SLC22A4
|
solute carrier family 22 (organic cation/zwitterion transporter), member 4 |
| chr6_+_31730773 | 0.25 |
ENST00000415669.2
ENST00000425424.1 |
SAPCD1
|
suppressor APC domain containing 1 |
| chr11_-_2292182 | 0.25 |
ENST00000331289.4
|
ASCL2
|
achaete-scute family bHLH transcription factor 2 |
| chr5_+_66124590 | 0.24 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr17_+_39421591 | 0.24 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr8_-_121824374 | 0.24 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr9_+_19408919 | 0.23 |
ENST00000380376.1
|
ACER2
|
alkaline ceramidase 2 |
| chr19_-_35626104 | 0.23 |
ENST00000310123.3
ENST00000392225.3 |
LGI4
|
leucine-rich repeat LGI family, member 4 |
| chr2_-_164592497 | 0.22 |
ENST00000333129.3
ENST00000409634.1 |
FIGN
|
fidgetin |
| chr19_+_56905024 | 0.22 |
ENST00000591172.1
ENST00000589888.1 ENST00000587979.1 ENST00000585659.1 ENST00000593109.1 |
ZNF582-AS1
|
ZNF582 antisense RNA 1 (head to head) |
| chr15_-_45670717 | 0.22 |
ENST00000558163.1
ENST00000558336.1 |
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr8_-_72268721 | 0.22 |
ENST00000419131.1
ENST00000388743.2 |
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr17_-_43339474 | 0.22 |
ENST00000331780.4
|
SPATA32
|
spermatogenesis associated 32 |
| chr20_-_52645231 | 0.22 |
ENST00000448484.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr10_+_1102303 | 0.21 |
ENST00000381329.1
|
WDR37
|
WD repeat domain 37 |
| chr11_+_62556596 | 0.21 |
ENST00000526546.1
|
TMEM179B
|
transmembrane protein 179B |
| chr12_+_6309963 | 0.21 |
ENST00000382515.2
|
CD9
|
CD9 molecule |
| chr10_-_73848531 | 0.21 |
ENST00000373109.2
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr19_+_18451439 | 0.20 |
ENST00000597431.2
|
PGPEP1
|
pyroglutamyl-peptidase I |
| chr17_-_43339453 | 0.20 |
ENST00000543122.1
|
SPATA32
|
spermatogenesis associated 32 |
| chr6_-_160147925 | 0.20 |
ENST00000535561.1
|
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr17_+_7239821 | 0.20 |
ENST00000158762.3
ENST00000570457.2 |
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr21_+_17791648 | 0.19 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_-_2461684 | 0.19 |
ENST00000378453.3
|
HES5
|
hes family bHLH transcription factor 5 |
| chr17_-_41132088 | 0.19 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr14_+_104710541 | 0.18 |
ENST00000419115.1
|
C14orf144
|
chromosome 14 open reading frame 144 |
| chr16_-_67427389 | 0.18 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr21_+_30503282 | 0.18 |
ENST00000399925.1
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr11_+_64004888 | 0.17 |
ENST00000541681.1
|
VEGFB
|
vascular endothelial growth factor B |
| chr5_-_169816638 | 0.17 |
ENST00000521859.1
ENST00000274629.4 |
KCNMB1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr16_+_83932684 | 0.17 |
ENST00000262430.4
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr8_-_29120604 | 0.16 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr2_-_61389168 | 0.16 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr17_+_68165657 | 0.15 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr15_-_48937982 | 0.15 |
ENST00000316623.5
|
FBN1
|
fibrillin 1 |
| chr4_+_103422499 | 0.15 |
ENST00000511926.1
ENST00000507079.1 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr15_-_45670924 | 0.15 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr17_-_7165662 | 0.15 |
ENST00000571881.2
ENST00000360325.7 |
CLDN7
|
claudin 7 |
| chr14_+_62331592 | 0.15 |
ENST00000554436.1
|
CTD-2277K2.1
|
CTD-2277K2.1 |
| chr3_-_128294929 | 0.15 |
ENST00000356020.2
|
C3orf27
|
chromosome 3 open reading frame 27 |
| chr16_+_103816 | 0.14 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr7_+_140103842 | 0.14 |
ENST00000495590.1
ENST00000275874.5 ENST00000537763.1 |
RAB19
|
RAB19, member RAS oncogene family |
| chr1_-_219615984 | 0.14 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr12_-_47226152 | 0.14 |
ENST00000546940.1
|
SLC38A4
|
solute carrier family 38, member 4 |
| chr8_-_120868078 | 0.14 |
ENST00000313655.4
|
DSCC1
|
DNA replication and sister chromatid cohesion 1 |
| chr16_-_103572 | 0.14 |
ENST00000293860.5
|
POLR3K
|
polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa |
| chr9_+_6758024 | 0.14 |
ENST00000442236.2
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr19_-_50316517 | 0.14 |
ENST00000313777.4
ENST00000445575.2 |
FUZ
|
fuzzy planar cell polarity protein |
| chr12_+_6309517 | 0.14 |
ENST00000382519.4
ENST00000009180.4 |
CD9
|
CD9 molecule |
| chrX_-_102565932 | 0.14 |
ENST00000372674.1
ENST00000372677.3 |
BEX2
|
brain expressed X-linked 2 |
| chr21_+_17791838 | 0.14 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr19_+_40877583 | 0.14 |
ENST00000596470.1
|
PLD3
|
phospholipase D family, member 3 |
| chr3_+_32280159 | 0.14 |
ENST00000458535.2
ENST00000307526.3 |
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr13_-_24471194 | 0.14 |
ENST00000382137.3
ENST00000382057.3 |
C1QTNF9B
|
C1q and tumor necrosis factor related protein 9B |
| chr3_+_54157480 | 0.14 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr2_-_120124258 | 0.13 |
ENST00000409877.1
ENST00000409523.1 ENST00000409466.2 ENST00000414534.1 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr2_+_74229812 | 0.13 |
ENST00000305799.7
|
TET3
|
tet methylcytosine dioxygenase 3 |
| chr20_-_44455976 | 0.13 |
ENST00000372555.3
|
TNNC2
|
troponin C type 2 (fast) |
| chr8_-_72268889 | 0.13 |
ENST00000388742.4
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr4_-_40632605 | 0.13 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr5_-_131630931 | 0.12 |
ENST00000431054.1
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr2_-_227050079 | 0.12 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chrX_-_33357558 | 0.12 |
ENST00000288447.4
|
DMD
|
dystrophin |
| chr15_+_57668695 | 0.12 |
ENST00000281282.5
|
CGNL1
|
cingulin-like 1 |
| chr1_-_85156417 | 0.12 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr2_+_233404429 | 0.12 |
ENST00000389494.3
ENST00000389492.3 |
CHRNG
|
cholinergic receptor, nicotinic, gamma (muscle) |
| chr17_+_48133459 | 0.12 |
ENST00000320031.8
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr19_+_18794470 | 0.12 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr19_-_50316489 | 0.12 |
ENST00000533418.1
|
FUZ
|
fuzzy planar cell polarity protein |
| chrX_-_111923145 | 0.11 |
ENST00000371968.3
ENST00000536453.1 |
LHFPL1
|
lipoma HMGIC fusion partner-like 1 |
| chr11_-_47470703 | 0.11 |
ENST00000298854.2
|
RAPSN
|
receptor-associated protein of the synapse |
| chr6_-_42713792 | 0.11 |
ENST00000372876.1
|
TBCC
|
tubulin folding cofactor C |
| chr22_+_22988816 | 0.11 |
ENST00000480559.1
ENST00000448514.1 |
GGTLC2
|
gamma-glutamyltransferase light chain 2 |
| chrX_+_154114635 | 0.11 |
ENST00000369446.2
|
F8A1
|
coagulation factor VIII-associated 1 |
| chr1_+_110026544 | 0.11 |
ENST00000369870.3
|
ATXN7L2
|
ataxin 7-like 2 |
| chr7_-_130598059 | 0.11 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chr3_-_24207039 | 0.11 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr17_-_42277203 | 0.11 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr19_-_7939319 | 0.11 |
ENST00000539422.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chr17_+_36508111 | 0.11 |
ENST00000331159.5
ENST00000577233.1 |
SOCS7
|
suppressor of cytokine signaling 7 |
| chr6_+_44184653 | 0.11 |
ENST00000573382.2
ENST00000576476.1 |
RP1-302G2.5
|
RP1-302G2.5 |
| chr17_-_56492989 | 0.11 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr5_+_50679506 | 0.11 |
ENST00000511384.1
|
ISL1
|
ISL LIM homeobox 1 |
| chr6_+_33378517 | 0.11 |
ENST00000428274.1
|
PHF1
|
PHD finger protein 1 |
| chr22_+_31489344 | 0.11 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr8_-_144691718 | 0.11 |
ENST00000377579.3
ENST00000433751.1 ENST00000220966.6 |
PYCRL
|
pyrroline-5-carboxylate reductase-like |
| chr17_+_4981535 | 0.10 |
ENST00000318833.3
|
ZFP3
|
ZFP3 zinc finger protein |
| chr1_+_226411319 | 0.10 |
ENST00000542034.1
ENST00000366810.5 |
MIXL1
|
Mix paired-like homeobox |
| chr10_-_73848764 | 0.10 |
ENST00000317376.4
ENST00000412663.1 |
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr15_-_94614049 | 0.10 |
ENST00000556447.1
ENST00000555772.1 |
CTD-3049M7.1
|
CTD-3049M7.1 |
| chr9_+_6757634 | 0.10 |
ENST00000543771.1
ENST00000401787.3 ENST00000381306.3 ENST00000381309.3 |
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr19_+_54371114 | 0.10 |
ENST00000448420.1
ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM
|
myeloid-associated differentiation marker |
| chr11_-_47470591 | 0.10 |
ENST00000524487.1
|
RAPSN
|
receptor-associated protein of the synapse |
| chr17_+_43299241 | 0.10 |
ENST00000328118.3
|
FMNL1
|
formin-like 1 |
| chr1_+_10509971 | 0.10 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr17_-_80009650 | 0.10 |
ENST00000310496.4
|
RFNG
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr16_-_30022293 | 0.09 |
ENST00000565273.1
ENST00000567332.2 ENST00000350119.4 |
DOC2A
|
double C2-like domains, alpha |
| chr17_+_47653178 | 0.09 |
ENST00000328741.5
|
NXPH3
|
neurexophilin 3 |
| chr3_-_87039662 | 0.09 |
ENST00000494229.1
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr1_-_207096529 | 0.09 |
ENST00000525793.1
ENST00000529560.1 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr1_-_161015752 | 0.09 |
ENST00000435396.1
ENST00000368021.3 |
USF1
|
upstream transcription factor 1 |
| chr15_+_75660431 | 0.09 |
ENST00000563278.1
|
RP11-817O13.8
|
RP11-817O13.8 |
| chrX_-_40005865 | 0.09 |
ENST00000412952.1
|
BCOR
|
BCL6 corepressor |
| chr9_-_35619539 | 0.09 |
ENST00000396757.1
|
CD72
|
CD72 molecule |
| chr17_-_46657473 | 0.09 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr17_-_79792909 | 0.09 |
ENST00000330261.4
ENST00000570394.1 |
PPP1R27
|
protein phosphatase 1, regulatory subunit 27 |
| chr2_-_61389240 | 0.09 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr12_-_70093235 | 0.09 |
ENST00000266661.4
|
BEST3
|
bestrophin 3 |
| chr5_-_59783882 | 0.09 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr19_+_16187085 | 0.09 |
ENST00000300933.4
|
TPM4
|
tropomyosin 4 |
| chr7_-_76255444 | 0.09 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr2_+_121493717 | 0.09 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chrX_+_86772787 | 0.09 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chrX_+_30671476 | 0.09 |
ENST00000378946.3
ENST00000378943.3 ENST00000378945.3 ENST00000427190.1 ENST00000378941.3 |
GK
|
glycerol kinase |
| chr11_-_118868682 | 0.09 |
ENST00000526453.1
|
RP11-110I1.12
|
RP11-110I1.12 |
| chr2_+_191045656 | 0.09 |
ENST00000443551.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr8_-_59572093 | 0.09 |
ENST00000427130.2
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr3_-_52719546 | 0.09 |
ENST00000439181.1
ENST00000449505.1 |
PBRM1
|
polybromo 1 |
| chr11_+_57365150 | 0.09 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr20_+_35202909 | 0.09 |
ENST00000558530.1
ENST00000558028.1 ENST00000560025.1 |
TGIF2-C20orf24
TGIF2
|
TGIF2-C20orf24 readthrough TGFB-induced factor homeobox 2 |
| chr12_-_57006882 | 0.09 |
ENST00000551996.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chrX_-_154688276 | 0.09 |
ENST00000369445.2
|
F8A3
|
coagulation factor VIII-associated 3 |
| chr21_+_30502806 | 0.09 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr1_-_223537475 | 0.09 |
ENST00000344029.6
ENST00000494793.2 ENST00000366878.4 ENST00000366877.3 |
SUSD4
|
sushi domain containing 4 |
| chr10_+_7860460 | 0.09 |
ENST00000344293.5
|
TAF3
|
TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa |
| chr17_+_47653471 | 0.09 |
ENST00000513748.1
|
NXPH3
|
neurexophilin 3 |
| chr9_+_19408999 | 0.09 |
ENST00000340967.2
|
ACER2
|
alkaline ceramidase 2 |
| chr14_+_68086515 | 0.09 |
ENST00000261783.3
|
ARG2
|
arginase 2 |
| chr11_-_30038490 | 0.08 |
ENST00000328224.6
|
KCNA4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr6_+_96463840 | 0.08 |
ENST00000302103.5
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase) |
| chr8_-_29120580 | 0.08 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr17_-_77925806 | 0.08 |
ENST00000574241.2
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr12_-_42632127 | 0.08 |
ENST00000555248.2
|
YAF2
|
YY1 associated factor 2 |
| chr8_-_37594944 | 0.08 |
ENST00000330539.1
|
RP11-863K10.7
|
Uncharacterized protein |
| chr15_-_43559055 | 0.08 |
ENST00000220420.5
ENST00000349114.4 |
TGM5
|
transglutaminase 5 |
| chrX_-_64254587 | 0.08 |
ENST00000337990.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr16_-_1968231 | 0.08 |
ENST00000443547.1
ENST00000293937.3 ENST00000454677.2 |
HS3ST6
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 6 |
| chr6_+_17281573 | 0.08 |
ENST00000379052.5
|
RBM24
|
RNA binding motif protein 24 |
| chr3_+_99833755 | 0.08 |
ENST00000489081.1
|
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr1_+_162351503 | 0.08 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr5_-_176936844 | 0.08 |
ENST00000510380.1
ENST00000510898.1 ENST00000357198.4 |
DOK3
|
docking protein 3 |
| chr12_+_121088291 | 0.08 |
ENST00000351200.2
|
CABP1
|
calcium binding protein 1 |
| chr19_-_42573650 | 0.08 |
ENST00000593562.1
|
GRIK5
|
glutamate receptor, ionotropic, kainate 5 |
| chr17_+_42925270 | 0.08 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr14_+_62584197 | 0.08 |
ENST00000334389.4
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
| chr9_+_100174344 | 0.08 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr11_-_76155618 | 0.08 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr2_-_30144432 | 0.08 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr1_+_38273818 | 0.08 |
ENST00000373042.4
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr4_+_184826418 | 0.08 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chr1_+_179851893 | 0.08 |
ENST00000531630.2
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chr1_-_161015663 | 0.08 |
ENST00000534633.1
|
USF1
|
upstream transcription factor 1 |
| chr9_-_79307096 | 0.08 |
ENST00000376717.2
ENST00000223609.6 ENST00000443509.2 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr16_+_31044413 | 0.08 |
ENST00000394998.1
|
STX4
|
syntaxin 4 |
| chr11_-_8954491 | 0.08 |
ENST00000526227.1
ENST00000525780.1 ENST00000326053.5 |
C11orf16
|
chromosome 11 open reading frame 16 |
| chr4_+_169418255 | 0.07 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr9_+_131683174 | 0.07 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr3_+_52719936 | 0.07 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chrX_+_129305623 | 0.07 |
ENST00000257017.4
|
RAB33A
|
RAB33A, member RAS oncogene family |
| chr11_+_71238313 | 0.07 |
ENST00000398536.4
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr5_+_118406796 | 0.07 |
ENST00000503802.1
|
DMXL1
|
Dmx-like 1 |
| chr12_-_27167233 | 0.07 |
ENST00000535819.1
ENST00000543803.1 ENST00000535423.1 ENST00000539741.1 ENST00000343028.4 ENST00000545600.1 ENST00000543088.1 |
TM7SF3
|
transmembrane 7 superfamily member 3 |
| chr11_-_123525648 | 0.07 |
ENST00000527836.1
|
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr5_+_102201687 | 0.07 |
ENST00000304400.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr10_-_128975273 | 0.07 |
ENST00000424811.2
|
FAM196A
|
family with sequence similarity 196, member A |
| chr19_-_46476791 | 0.07 |
ENST00000263257.5
|
NOVA2
|
neuro-oncological ventral antigen 2 |
| chr2_+_48541776 | 0.07 |
ENST00000413569.1
ENST00000340553.3 |
FOXN2
|
forkhead box N2 |
| chr3_+_50192833 | 0.07 |
ENST00000426511.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr1_-_54872059 | 0.07 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr9_+_118950325 | 0.07 |
ENST00000534838.1
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr19_-_48894104 | 0.07 |
ENST00000597017.1
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr17_-_39623681 | 0.07 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chrX_-_70473957 | 0.07 |
ENST00000373984.3
ENST00000314425.5 ENST00000373982.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chrX_-_151619746 | 0.07 |
ENST00000370314.4
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr5_+_56111361 | 0.07 |
ENST00000399503.3
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr12_-_54653313 | 0.07 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr5_-_159739532 | 0.07 |
ENST00000520748.1
ENST00000393977.3 ENST00000257536.7 |
CCNJL
|
cyclin J-like |
| chr10_+_104221137 | 0.07 |
ENST00000366277.2
ENST00000238936.4 ENST00000369931.3 |
TMEM180
|
transmembrane protein 180 |
| chr10_+_1102721 | 0.07 |
ENST00000263150.4
|
WDR37
|
WD repeat domain 37 |
| chr10_+_103348031 | 0.07 |
ENST00000370151.4
ENST00000370147.1 ENST00000370148.2 |
DPCD
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr8_+_145065705 | 0.07 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chrX_-_11445856 | 0.07 |
ENST00000380736.1
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chrX_-_43832711 | 0.07 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chrX_+_107334983 | 0.07 |
ENST00000457035.1
ENST00000545696.1 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chr14_+_24600484 | 0.07 |
ENST00000267426.5
|
FITM1
|
fat storage-inducing transmembrane protein 1 |
| chr8_-_135522425 | 0.07 |
ENST00000521673.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr15_-_75748115 | 0.07 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr2_+_120124497 | 0.07 |
ENST00000355857.3
ENST00000535617.1 ENST00000535757.1 ENST00000409094.1 ENST00000311521.4 |
DBI
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr5_-_159739483 | 0.07 |
ENST00000519673.1
ENST00000541762.1 |
CCNJL
|
cyclin J-like |
| chr9_+_100174232 | 0.06 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MYOD1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.4 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.3 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.3 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.3 | GO:0090299 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 | 0.2 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.1 | 0.3 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.5 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 0.2 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.0 | 0.1 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.3 | GO:0031179 | peptide amidation(GO:0001519) peptide modification(GO:0031179) |
| 0.0 | 0.2 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.2 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.4 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.2 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.2 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.2 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:2000744 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0060381 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:0035482 | gastric motility(GO:0035482) gastric emptying(GO:0035483) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:1902566 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.0 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.0 | 0.0 | GO:0002585 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.0 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.0 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.1 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.0 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.1 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.1 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.0 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.0 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.3 | GO:1902603 | carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.0 | GO:0035284 | rhombomere 3 development(GO:0021569) rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.1 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015067 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.1 | 0.3 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.1 | 0.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.3 | GO:0015199 | secondary active organic cation transmembrane transporter activity(GO:0008513) amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.2 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.0 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.0 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.0 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |