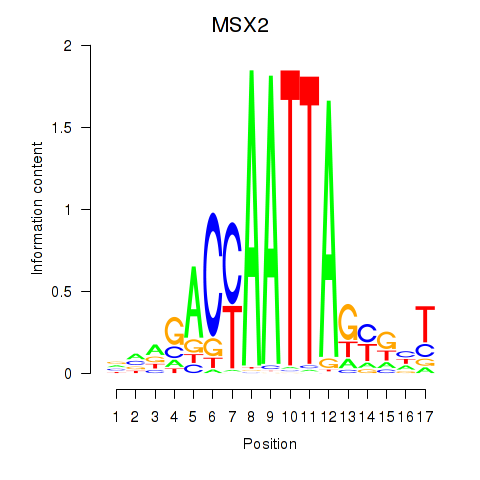
|
chr14_-_88200641
Show fit
|
0.24 |
ENST00000556168.1
|
RP11-1152H15.1
|
RP11-1152H15.1
|
|
chr1_+_152943122
Show fit
|
0.19 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4
|
|
chr3_-_180397256
Show fit
|
0.11 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39
|
|
chr12_+_64798826
Show fit
|
0.10 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA
|
|
chr1_+_81001398
Show fit
|
0.10 |
ENST00000418041.1
ENST00000443104.1
|
RP5-887A10.1
|
RP5-887A10.1
|
|
chr20_-_50418972
Show fit
|
0.09 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4
|
|
chr3_-_112127981
Show fit
|
0.08 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1
|
|
chr7_-_2883650
Show fit
|
0.08 |
ENST00000544127.1
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12
|
|
chr16_-_18923035
Show fit
|
0.08 |
ENST00000563836.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase
|
|
chr6_+_130339710
Show fit
|
0.08 |
ENST00000526087.1
ENST00000533560.1
ENST00000361794.2
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr12_-_9102224
Show fit
|
0.08 |
ENST00000543845.1
ENST00000544245.1
|
M6PR
|
mannose-6-phosphate receptor (cation dependent)
|
|
chr8_-_117886563
Show fit
|
0.07 |
ENST00000519837.1
ENST00000522699.1
|
RAD21
|
RAD21 homolog (S. pombe)
|
|
chr1_-_151762900
Show fit
|
0.07 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing
|
|
chr16_+_58783542
Show fit
|
0.07 |
ENST00000500117.1
ENST00000565722.1
|
RP11-410D17.2
|
RP11-410D17.2
|
|
chr10_-_99205607
Show fit
|
0.06 |
ENST00000477692.2
ENST00000485122.2
ENST00000370886.5
ENST00000370885.4
ENST00000370902.3
ENST00000370884.5
|
EXOSC1
|
exosome component 1
|
|
chr3_-_120400960
Show fit
|
0.06 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr3_-_141747950
Show fit
|
0.06 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr11_+_17298543
Show fit
|
0.06 |
ENST00000533926.1
|
NUCB2
|
nucleobindin 2
|
|
chr18_+_63273310
Show fit
|
0.06 |
ENST00000579862.1
|
RP11-775G23.1
|
RP11-775G23.1
|
|
chr4_-_69434245
Show fit
|
0.06 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17
|
|
chr7_+_99717230
Show fit
|
0.05 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4
|
|
chr1_+_44115814
Show fit
|
0.05 |
ENST00000372396.3
|
KDM4A
|
lysine (K)-specific demethylase 4A
|
|
chr19_+_41869894
Show fit
|
0.05 |
ENST00000413014.2
|
TMEM91
|
transmembrane protein 91
|
|
chr6_-_167040731
Show fit
|
0.05 |
ENST00000265678.4
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr10_+_99205894
Show fit
|
0.05 |
ENST00000370854.3
ENST00000393760.1
ENST00000414567.1
ENST00000370846.4
|
ZDHHC16
|
zinc finger, DHHC-type containing 16
|
|
chr7_-_2883928
Show fit
|
0.05 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12
|
|
chr10_+_99205959
Show fit
|
0.04 |
ENST00000352634.4
ENST00000353979.3
ENST00000370842.2
ENST00000345745.5
|
ZDHHC16
|
zinc finger, DHHC-type containing 16
|
|
chr17_-_43045439
Show fit
|
0.04 |
ENST00000253407.3
|
C1QL1
|
complement component 1, q subcomponent-like 1
|
|
chrX_+_77154935
Show fit
|
0.04 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb
|
|
chr12_-_120315074
Show fit
|
0.04 |
ENST00000261833.7
ENST00000392521.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chr3_-_120003941
Show fit
|
0.04 |
ENST00000464295.1
|
GPR156
|
G protein-coupled receptor 156
|
|
chr12_-_95397442
Show fit
|
0.04 |
ENST00000547157.1
ENST00000547986.1
ENST00000327772.2
|
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12
|
|
chr1_-_151762943
Show fit
|
0.04 |
ENST00000368825.3
ENST00000368823.1
ENST00000458431.2
ENST00000368827.6
ENST00000368824.3
|
TDRKH
|
tudor and KH domain containing
|
|
chr12_-_9102549
Show fit
|
0.04 |
ENST00000000412.3
|
M6PR
|
mannose-6-phosphate receptor (cation dependent)
|
|
chr6_-_116866773
Show fit
|
0.03 |
ENST00000368602.3
|
TRAPPC3L
|
trafficking protein particle complex 3-like
|
|
chr12_-_57039739
Show fit
|
0.03 |
ENST00000552959.1
ENST00000551020.1
ENST00000553007.2
ENST00000552919.1
ENST00000552104.1
ENST00000262030.3
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide
|
|
chr3_+_113251143
Show fit
|
0.03 |
ENST00000264852.4
ENST00000393830.3
|
SIDT1
|
SID1 transmembrane family, member 1
|
|
chr15_-_37393406
Show fit
|
0.03 |
ENST00000338564.5
ENST00000558313.1
ENST00000340545.5
|
MEIS2
|
Meis homeobox 2
|
|
chr1_+_27158483
Show fit
|
0.03 |
ENST00000374141.2
|
ZDHHC18
|
zinc finger, DHHC-type containing 18
|
|
chr20_-_54967187
Show fit
|
0.03 |
ENST00000422322.1
ENST00000371356.2
ENST00000451915.1
ENST00000347343.2
ENST00000395911.1
ENST00000395907.1
ENST00000441357.1
ENST00000456249.1
ENST00000420474.1
ENST00000395909.4
ENST00000395914.1
ENST00000312783.6
ENST00000395915.3
ENST00000395913.3
|
AURKA
|
aurora kinase A
|
|
chr11_+_108535752
Show fit
|
0.03 |
ENST00000322536.3
|
DDX10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10
|
|
chr1_-_151965048
Show fit
|
0.03 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10
|
|
chr5_-_31532238
Show fit
|
0.03 |
ENST00000507438.1
|
DROSHA
|
drosha, ribonuclease type III
|
|
chr5_+_95998070
Show fit
|
0.03 |
ENST00000421689.2
ENST00000510756.1
ENST00000512620.1
|
CAST
|
calpastatin
|
|
chr1_-_47779762
Show fit
|
0.03 |
ENST00000371877.3
ENST00000360380.3
ENST00000337817.5
ENST00000447475.2
|
STIL
|
SCL/TAL1 interrupting locus
|
|
chr5_-_132200477
Show fit
|
0.03 |
ENST00000296875.2
|
GDF9
|
growth differentiation factor 9
|
|
chr5_+_150639360
Show fit
|
0.03 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator
|
|
chr17_+_67759813
Show fit
|
0.03 |
ENST00000587241.1
|
AC003051.1
|
AC003051.1
|
|
chr20_+_54967663
Show fit
|
0.03 |
ENST00000452950.1
|
CSTF1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa
|
|
chr1_-_151431674
Show fit
|
0.03 |
ENST00000531094.1
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr6_-_167040693
Show fit
|
0.03 |
ENST00000366863.2
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr1_-_68698197
Show fit
|
0.03 |
ENST00000370973.2
ENST00000370971.1
|
WLS
|
wntless Wnt ligand secretion mediator
|
|
chr9_+_34646624
Show fit
|
0.03 |
ENST00000450095.2
ENST00000556278.1
|
GALT
GALT
|
galactose-1-phosphate uridylyltransferase
Uncharacterized protein
|
|
chr20_-_50418947
Show fit
|
0.03 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4
|
|
chr14_-_23504432
Show fit
|
0.03 |
ENST00000425762.2
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5
|
|
chr20_-_50722183
Show fit
|
0.03 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein
|
|
chr11_-_124981475
Show fit
|
0.02 |
ENST00000532156.1
ENST00000532407.1
ENST00000279968.4
ENST00000527766.1
ENST00000529583.1
ENST00000524373.1
ENST00000527271.1
ENST00000526175.1
ENST00000529609.1
ENST00000533273.1
ENST00000531909.1
ENST00000529530.1
|
TMEM218
|
transmembrane protein 218
|
|
chr7_+_100183927
Show fit
|
0.02 |
ENST00000241071.6
ENST00000360609.2
|
FBXO24
|
F-box protein 24
|
|
chr2_-_200335983
Show fit
|
0.02 |
ENST00000457245.1
|
SATB2
|
SATB homeobox 2
|
|
chr9_+_34646651
Show fit
|
0.02 |
ENST00000378842.3
|
GALT
|
galactose-1-phosphate uridylyltransferase
|
|
chr12_+_54718904
Show fit
|
0.02 |
ENST00000262061.2
ENST00000549043.1
ENST00000552218.1
ENST00000553231.1
ENST00000552362.1
ENST00000455864.2
ENST00000416254.2
ENST00000549116.1
ENST00000551779.1
|
COPZ1
|
coatomer protein complex, subunit zeta 1
|
|
chr5_+_95997885
Show fit
|
0.02 |
ENST00000511097.2
|
CAST
|
calpastatin
|
|
chr17_-_12920907
Show fit
|
0.02 |
ENST00000609757.1
ENST00000581499.2
ENST00000580504.1
|
ELAC2
|
elaC ribonuclease Z 2
|
|
chr16_+_69345243
Show fit
|
0.02 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae)
|
|
chr1_-_36916066
Show fit
|
0.02 |
ENST00000315643.9
|
OSCP1
|
organic solute carrier partner 1
|
|
chrX_-_48931648
Show fit
|
0.02 |
ENST00000376386.3
ENST00000376390.4
|
PRAF2
|
PRA1 domain family, member 2
|
|
chr17_+_12569306
Show fit
|
0.02 |
ENST00000425538.1
|
MYOCD
|
myocardin
|
|
chr11_-_71823715
Show fit
|
0.02 |
ENST00000545944.1
ENST00000502597.2
|
ANAPC15
|
anaphase promoting complex subunit 15
|
|
chr6_-_111927062
Show fit
|
0.02 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr1_-_151431647
Show fit
|
0.02 |
ENST00000368863.2
ENST00000409503.1
ENST00000491586.1
ENST00000533351.1
ENST00000540984.1
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr5_-_31532160
Show fit
|
0.02 |
ENST00000511367.2
ENST00000513349.1
|
DROSHA
|
drosha, ribonuclease type III
|
|
chr17_-_9929581
Show fit
|
0.02 |
ENST00000437099.2
ENST00000396115.2
|
GAS7
|
growth arrest-specific 7
|
|
chr4_-_69536346
Show fit
|
0.02 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15
|
|
chr2_+_150187020
Show fit
|
0.02 |
ENST00000334166.4
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr1_-_158173659
Show fit
|
0.02 |
ENST00000415019.1
|
RP11-404O13.5
|
RP11-404O13.5
|
|
chr7_-_99716940
Show fit
|
0.02 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa
|
|
chr14_-_23504337
Show fit
|
0.02 |
ENST00000361611.6
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5
|
|
chr7_+_134331550
Show fit
|
0.02 |
ENST00000344924.3
ENST00000418040.1
ENST00000393132.2
|
BPGM
|
2,3-bisphosphoglycerate mutase
|
|
chr6_+_30687978
Show fit
|
0.02 |
ENST00000327892.8
ENST00000435534.1
|
TUBB
|
tubulin, beta class I
|
|
chr17_+_12569472
Show fit
|
0.01 |
ENST00000343344.4
|
MYOCD
|
myocardin
|
|
chrX_+_70752917
Show fit
|
0.01 |
ENST00000373719.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase
|
|
chr11_+_17298342
Show fit
|
0.01 |
ENST00000530964.1
|
NUCB2
|
nucleobindin 2
|
|
chr1_+_151735431
Show fit
|
0.01 |
ENST00000321531.5
ENST00000315067.8
|
OAZ3
|
ornithine decarboxylase antizyme 3
|
|
chr22_-_30960876
Show fit
|
0.01 |
ENST00000401975.1
ENST00000428682.1
ENST00000423299.1
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1
|
|
chr3_+_186353756
Show fit
|
0.01 |
ENST00000431018.1
ENST00000450521.1
ENST00000539949.1
|
FETUB
|
fetuin B
|
|
chr9_+_74729511
Show fit
|
0.01 |
ENST00000545168.1
|
GDA
|
guanine deaminase
|
|
chr20_+_30467600
Show fit
|
0.01 |
ENST00000375934.4
ENST00000375922.4
|
TTLL9
|
tubulin tyrosine ligase-like family, member 9
|
|
chr4_-_109684120
Show fit
|
0.01 |
ENST00000512646.1
ENST00000411864.2
ENST00000296486.3
ENST00000510706.1
|
ETNPPL
|
ethanolamine-phosphate phospho-lyase
|
|
chr1_-_23694794
Show fit
|
0.01 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436
|
|
chr1_-_94050668
Show fit
|
0.01 |
ENST00000539242.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chrX_+_70752945
Show fit
|
0.01 |
ENST00000373701.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase
|
|
chr6_-_31830655
Show fit
|
0.01 |
ENST00000375631.4
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr7_+_130794846
Show fit
|
0.01 |
ENST00000421797.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr15_+_80351910
Show fit
|
0.01 |
ENST00000261749.6
ENST00000561060.1
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr3_-_48672859
Show fit
|
0.01 |
ENST00000395550.2
ENST00000455886.2
ENST00000431739.1
ENST00000426599.1
ENST00000383733.3
ENST00000420764.2
ENST00000337000.8
|
SLC26A6
|
solute carrier family 26 (anion exchanger), member 6
|
|
chr20_+_49126881
Show fit
|
0.01 |
ENST00000371621.3
ENST00000541713.1
|
PTPN1
|
protein tyrosine phosphatase, non-receptor type 1
|
|
chr6_+_160183492
Show fit
|
0.01 |
ENST00000541436.1
|
ACAT2
|
acetyl-CoA acetyltransferase 2
|
|
chr11_-_71823796
Show fit
|
0.01 |
ENST00000545680.1
ENST00000543587.1
ENST00000538393.1
ENST00000535234.1
ENST00000227618.4
ENST00000535503.1
|
ANAPC15
|
anaphase promoting complex subunit 15
|
|
chr4_-_138453606
Show fit
|
0.01 |
ENST00000412923.2
ENST00000344876.4
ENST00000507846.1
ENST00000510305.1
|
PCDH18
|
protocadherin 18
|
|
chr19_+_17378278
Show fit
|
0.01 |
ENST00000596335.1
ENST00000601436.1
ENST00000595632.1
|
BABAM1
|
BRISC and BRCA1 A complex member 1
|
|
chr1_-_21377383
Show fit
|
0.01 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr8_+_38831683
Show fit
|
0.01 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4
|
|
chr1_-_68698222
Show fit
|
0.01 |
ENST00000370976.3
ENST00000354777.2
ENST00000262348.4
ENST00000540432.1
|
WLS
|
wntless Wnt ligand secretion mediator
|
|
chr10_-_56561022
Show fit
|
0.01 |
ENST00000373965.2
ENST00000414778.1
ENST00000395438.1
ENST00000409834.1
ENST00000395445.1
ENST00000395446.1
ENST00000395442.1
ENST00000395440.1
ENST00000395432.2
ENST00000361849.3
ENST00000395433.1
ENST00000320301.6
ENST00000395430.1
ENST00000437009.1
|
PCDH15
|
protocadherin-related 15
|
|
chr3_-_114866084
Show fit
|
0.01 |
ENST00000357258.3
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr13_-_103389159
Show fit
|
0.01 |
ENST00000322527.2
|
CCDC168
|
coiled-coil domain containing 168
|
|
chr1_-_151431909
Show fit
|
0.00 |
ENST00000361398.3
ENST00000271715.2
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr2_-_98280383
Show fit
|
0.00 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast)
|
|
chr20_+_56964169
Show fit
|
0.00 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr9_+_80912059
Show fit
|
0.00 |
ENST00000347159.2
ENST00000376588.3
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chr11_-_71823266
Show fit
|
0.00 |
ENST00000538919.1
ENST00000539395.1
ENST00000542531.1
|
ANAPC15
|
anaphase promoting complex subunit 15
|
|
chr1_-_8877692
Show fit
|
0.00 |
ENST00000400908.2
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats
|
|
chr4_-_171012844
Show fit
|
0.00 |
ENST00000502392.1
|
AADAT
|
aminoadipate aminotransferase
|
|
chr8_+_1993173
Show fit
|
0.00 |
ENST00000523438.1
|
MYOM2
|
myomesin 2
|
|
chr2_+_187371440
Show fit
|
0.00 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr11_+_108535849
Show fit
|
0.00 |
ENST00000526794.1
|
DDX10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10
|
|
chr11_+_33061543
Show fit
|
0.00 |
ENST00000432887.1
ENST00000528898.1
ENST00000531632.2
|
TCP11L1
|
t-complex 11, testis-specific-like 1
|
|
chr9_-_5339873
Show fit
|
0.00 |
ENST00000223862.1
ENST00000223858.4
|
RLN1
|
relaxin 1
|
|
chr8_+_1993152
Show fit
|
0.00 |
ENST00000262113.4
|
MYOM2
|
myomesin 2
|
|
chr19_+_11485333
Show fit
|
0.00 |
ENST00000312423.2
|
SWSAP1
|
SWIM-type zinc finger 7 associated protein 1
|