Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
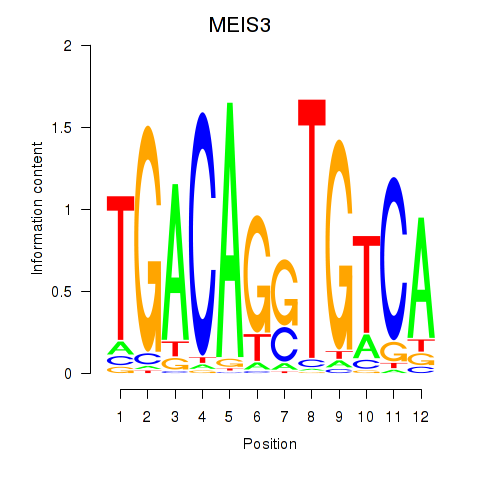
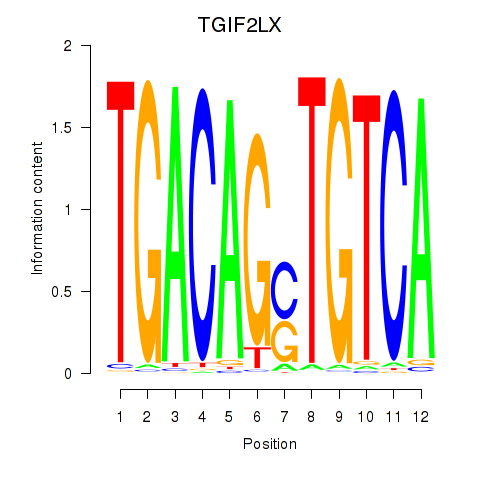
Results for MEIS3_TGIF2LX
Z-value: 0.67
Transcription factors associated with MEIS3_TGIF2LX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEIS3
|
ENSG00000105419.13 | Meis homeobox 3 |
|
TGIF2LX
|
ENSG00000153779.8 | TGFB induced factor homeobox 2 like X-linked |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEIS3 | hg19_v2_chr19_-_47922750_47922795 | 0.21 | 6.8e-01 | Click! |
Activity profile of MEIS3_TGIF2LX motif
Sorted Z-values of MEIS3_TGIF2LX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_47655686 | 1.09 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr1_-_153013588 | 0.80 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chr11_+_18287721 | 0.59 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr11_+_18287801 | 0.58 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr7_+_76139741 | 0.56 |
ENST00000334348.3
ENST00000419923.2 ENST00000448265.3 ENST00000443097.2 |
UPK3B
|
uroplakin 3B |
| chr14_+_97925151 | 0.40 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr19_+_1000418 | 0.38 |
ENST00000234389.3
|
GRIN3B
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B |
| chr11_+_102188224 | 0.27 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr7_+_76139833 | 0.27 |
ENST00000257632.5
|
UPK3B
|
uroplakin 3B |
| chr19_-_51531210 | 0.26 |
ENST00000391804.3
|
KLK11
|
kallikrein-related peptidase 11 |
| chr11_-_615570 | 0.26 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr3_-_49726486 | 0.26 |
ENST00000449682.2
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr7_+_134331550 | 0.24 |
ENST00000344924.3
ENST00000418040.1 ENST00000393132.2 |
BPGM
|
2,3-bisphosphoglycerate mutase |
| chr19_-_9938480 | 0.23 |
ENST00000585379.1
|
FBXL12
|
F-box and leucine-rich repeat protein 12 |
| chr11_+_102188272 | 0.23 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr2_+_38177575 | 0.22 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr18_+_68002675 | 0.21 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr21_-_43735446 | 0.20 |
ENST00000398431.2
|
TFF3
|
trefoil factor 3 (intestinal) |
| chr15_-_74284558 | 0.19 |
ENST00000359750.4
ENST00000541638.1 ENST00000562453.1 |
STOML1
|
stomatin (EPB72)-like 1 |
| chr2_+_97202480 | 0.19 |
ENST00000357485.3
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr17_-_1395954 | 0.19 |
ENST00000359786.5
|
MYO1C
|
myosin IC |
| chr19_+_6361754 | 0.18 |
ENST00000597326.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr7_+_76139925 | 0.18 |
ENST00000394849.1
|
UPK3B
|
uroplakin 3B |
| chr19_-_51531272 | 0.18 |
ENST00000319720.7
|
KLK11
|
kallikrein-related peptidase 11 |
| chr17_+_18380051 | 0.17 |
ENST00000581545.1
ENST00000582333.1 ENST00000328114.6 ENST00000412421.2 ENST00000583322.1 ENST00000584941.1 |
LGALS9C
|
lectin, galactoside-binding, soluble, 9C |
| chr1_+_79115503 | 0.17 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr19_-_10227503 | 0.16 |
ENST00000593054.1
|
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
| chr17_-_79533608 | 0.16 |
ENST00000572760.1
ENST00000573876.1 |
NPLOC4
|
nuclear protein localization 4 homolog (S. cerevisiae) |
| chrX_+_148622513 | 0.16 |
ENST00000393985.3
ENST00000423421.1 ENST00000423540.2 ENST00000434353.2 ENST00000514208.1 |
CXorf40A
|
chromosome X open reading frame 40A |
| chr10_+_99349450 | 0.16 |
ENST00000370640.3
|
C10orf62
|
chromosome 10 open reading frame 62 |
| chr2_+_26785409 | 0.16 |
ENST00000329615.3
ENST00000409392.1 |
C2orf70
|
chromosome 2 open reading frame 70 |
| chr12_+_113682066 | 0.16 |
ENST00000392569.4
ENST00000552542.1 |
TPCN1
|
two pore segment channel 1 |
| chr2_+_24150180 | 0.15 |
ENST00000404924.1
|
UBXN2A
|
UBX domain protein 2A |
| chr6_+_134273321 | 0.15 |
ENST00000457715.1
|
TBPL1
|
TBP-like 1 |
| chr19_+_18530184 | 0.15 |
ENST00000601357.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr6_+_134273300 | 0.15 |
ENST00000416965.1
|
TBPL1
|
TBP-like 1 |
| chr2_+_90248739 | 0.15 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr22_+_46731596 | 0.14 |
ENST00000381019.3
|
TRMU
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr17_-_6917755 | 0.14 |
ENST00000593646.1
|
AC040977.1
|
Uncharacterized protein |
| chr18_+_54318893 | 0.14 |
ENST00000593058.1
|
WDR7
|
WD repeat domain 7 |
| chr2_+_149402009 | 0.13 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr3_+_49726932 | 0.13 |
ENST00000327697.6
ENST00000432042.1 ENST00000454491.1 |
RNF123
|
ring finger protein 123 |
| chr11_-_842509 | 0.13 |
ENST00000322028.4
|
POLR2L
|
polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa |
| chr12_+_121647868 | 0.13 |
ENST00000359949.7
ENST00000541532.1 ENST00000543171.1 ENST00000538701.1 |
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr4_-_18023350 | 0.12 |
ENST00000539056.1
ENST00000382226.5 ENST00000326877.4 |
LCORL
|
ligand dependent nuclear receptor corepressor-like |
| chrX_-_23926004 | 0.12 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr15_-_74284613 | 0.12 |
ENST00000316911.6
ENST00000564777.1 ENST00000566081.1 ENST00000316900.5 |
STOML1
|
stomatin (EPB72)-like 1 |
| chr2_+_181988560 | 0.12 |
ENST00000424170.1
ENST00000435411.1 |
AC104820.2
|
AC104820.2 |
| chr9_-_22009241 | 0.12 |
ENST00000380142.4
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr21_-_46954529 | 0.11 |
ENST00000485649.2
|
SLC19A1
|
solute carrier family 19 (folate transporter), member 1 |
| chr9_-_90589402 | 0.11 |
ENST00000375871.4
ENST00000605159.1 ENST00000336654.5 |
CDK20
|
cyclin-dependent kinase 20 |
| chr19_-_51529849 | 0.11 |
ENST00000600362.1
ENST00000453757.3 ENST00000601671.1 |
KLK11
|
kallikrein-related peptidase 11 |
| chr20_+_35504522 | 0.11 |
ENST00000602922.1
ENST00000217320.3 |
TLDC2
|
TBC/LysM-associated domain containing 2 |
| chr14_+_21492331 | 0.10 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr9_+_34990219 | 0.10 |
ENST00000541010.1
ENST00000454002.2 ENST00000545841.1 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr19_-_51530916 | 0.10 |
ENST00000594768.1
|
KLK11
|
kallikrein-related peptidase 11 |
| chr8_-_61880248 | 0.10 |
ENST00000525556.1
|
AC022182.3
|
AC022182.3 |
| chr19_-_5680202 | 0.10 |
ENST00000590389.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr16_+_30212378 | 0.10 |
ENST00000569485.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr7_+_2599056 | 0.10 |
ENST00000325979.7
ENST00000423395.1 |
IQCE
|
IQ motif containing E |
| chr4_-_187517928 | 0.10 |
ENST00000512772.1
|
FAT1
|
FAT atypical cadherin 1 |
| chr19_+_6361795 | 0.10 |
ENST00000596149.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr17_+_7210294 | 0.09 |
ENST00000336452.7
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr19_-_5680231 | 0.09 |
ENST00000587950.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr19_-_58609570 | 0.09 |
ENST00000600845.1
ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18
|
zinc finger and SCAN domain containing 18 |
| chr3_-_47023455 | 0.09 |
ENST00000446836.1
ENST00000425441.1 |
CCDC12
|
coiled-coil domain containing 12 |
| chr12_+_652294 | 0.09 |
ENST00000322843.3
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr17_-_39781054 | 0.09 |
ENST00000463128.1
|
KRT17
|
keratin 17 |
| chr17_-_39780819 | 0.09 |
ENST00000311208.8
|
KRT17
|
keratin 17 |
| chr2_-_73460334 | 0.08 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chrX_+_153775821 | 0.08 |
ENST00000263518.6
ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr17_-_39780634 | 0.08 |
ENST00000577817.2
|
KRT17
|
keratin 17 |
| chr2_+_97203082 | 0.08 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr1_+_154975258 | 0.08 |
ENST00000417934.2
|
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr16_-_69418553 | 0.08 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr12_+_121647962 | 0.08 |
ENST00000542067.1
|
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr9_-_95055923 | 0.08 |
ENST00000430417.1
|
IARS
|
isoleucyl-tRNA synthetase |
| chrX_+_47420516 | 0.08 |
ENST00000377045.4
ENST00000290277.6 ENST00000377039.2 |
ARAF
|
v-raf murine sarcoma 3611 viral oncogene homolog |
| chr3_-_194188956 | 0.08 |
ENST00000256031.4
ENST00000446356.1 |
ATP13A3
|
ATPase type 13A3 |
| chr4_+_69313145 | 0.08 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr13_+_76413852 | 0.08 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr17_+_6916957 | 0.08 |
ENST00000547302.2
|
RNASEK-C17orf49
|
RNASEK-C17orf49 readthrough |
| chr5_+_166711804 | 0.07 |
ENST00000518659.1
ENST00000545108.1 |
TENM2
|
teneurin transmembrane protein 2 |
| chr11_-_26743546 | 0.07 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr8_+_56014949 | 0.07 |
ENST00000327381.6
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4 |
| chr8_+_23145594 | 0.07 |
ENST00000519952.1
ENST00000518840.1 |
R3HCC1
|
R3H domain and coiled-coil containing 1 |
| chr11_+_64073699 | 0.07 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr12_-_56615485 | 0.07 |
ENST00000549038.1
ENST00000552244.1 |
RNF41
|
ring finger protein 41 |
| chr10_+_97733786 | 0.07 |
ENST00000371198.2
|
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr11_-_2170786 | 0.07 |
ENST00000300632.5
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr2_-_179914760 | 0.07 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr1_+_46379254 | 0.07 |
ENST00000372008.2
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr8_+_63161550 | 0.07 |
ENST00000328472.5
|
NKAIN3
|
Na+/K+ transporting ATPase interacting 3 |
| chr15_+_65337708 | 0.07 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr12_-_53298841 | 0.07 |
ENST00000293308.6
|
KRT8
|
keratin 8 |
| chr1_-_169680745 | 0.06 |
ENST00000236147.4
|
SELL
|
selectin L |
| chr4_-_100871506 | 0.06 |
ENST00000296417.5
|
H2AFZ
|
H2A histone family, member Z |
| chr2_+_149402553 | 0.06 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr11_+_61447845 | 0.06 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr11_-_82745238 | 0.06 |
ENST00000531021.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr6_+_53948328 | 0.06 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr6_+_34204642 | 0.06 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chrX_-_153775760 | 0.06 |
ENST00000440967.1
ENST00000393564.2 ENST00000369620.2 |
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr1_+_154975110 | 0.06 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr11_-_72145669 | 0.06 |
ENST00000543042.1
ENST00000294053.3 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr21_+_30503282 | 0.06 |
ENST00000399925.1
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr6_+_53948221 | 0.06 |
ENST00000460844.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr4_+_78783674 | 0.06 |
ENST00000315567.8
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr11_-_65325664 | 0.06 |
ENST00000301873.5
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr13_+_78109804 | 0.06 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr16_+_56899114 | 0.06 |
ENST00000566786.1
ENST00000438926.2 ENST00000563236.1 ENST00000262502.5 |
SLC12A3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr19_-_40791302 | 0.06 |
ENST00000392038.2
ENST00000578123.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr9_-_113800705 | 0.06 |
ENST00000441240.1
|
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr1_-_16539094 | 0.05 |
ENST00000270747.3
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr11_-_72070206 | 0.05 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr22_+_21369316 | 0.05 |
ENST00000413302.2
ENST00000402329.3 ENST00000336296.2 ENST00000401443.1 ENST00000443995.3 |
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr4_-_44450814 | 0.05 |
ENST00000360029.3
|
KCTD8
|
potassium channel tetramerization domain containing 8 |
| chr21_+_30502806 | 0.05 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr13_+_96085847 | 0.05 |
ENST00000376873.3
|
CLDN10
|
claudin 10 |
| chr18_+_55018044 | 0.05 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr5_-_16738451 | 0.05 |
ENST00000274203.9
ENST00000515803.1 |
MYO10
|
myosin X |
| chrX_-_48326764 | 0.05 |
ENST00000413668.1
ENST00000441948.1 |
SLC38A5
|
solute carrier family 38, member 5 |
| chr5_+_142286887 | 0.05 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr17_-_33446820 | 0.05 |
ENST00000592577.1
ENST00000590016.1 ENST00000345365.6 ENST00000360276.3 ENST00000357906.3 |
RAD51D
|
RAD51 paralog D |
| chr22_+_39052632 | 0.05 |
ENST00000411557.1
ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1
|
chibby homolog 1 (Drosophila) |
| chr11_-_59612969 | 0.05 |
ENST00000541311.1
ENST00000257248.2 |
GIF
|
gastric intrinsic factor (vitamin B synthesis) |
| chr12_+_95611536 | 0.05 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr12_-_49412525 | 0.05 |
ENST00000551121.1
ENST00000552212.1 ENST00000548605.1 ENST00000548950.1 ENST00000547125.1 |
PRKAG1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr9_-_37592561 | 0.05 |
ENST00000544379.1
ENST00000377773.5 ENST00000401811.3 ENST00000321301.6 |
TOMM5
|
translocase of outer mitochondrial membrane 5 homolog (yeast) |
| chr10_+_104535994 | 0.04 |
ENST00000369889.4
|
WBP1L
|
WW domain binding protein 1-like |
| chr2_+_173724771 | 0.04 |
ENST00000538974.1
ENST00000540783.1 |
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr16_+_20817761 | 0.04 |
ENST00000568046.1
ENST00000261377.6 |
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chrX_+_153775869 | 0.04 |
ENST00000424839.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr17_-_5015129 | 0.04 |
ENST00000575898.1
ENST00000416429.2 |
ZNF232
|
zinc finger protein 232 |
| chr5_-_72861175 | 0.04 |
ENST00000504641.1
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr19_+_5623186 | 0.04 |
ENST00000538656.1
|
SAFB
|
scaffold attachment factor B |
| chr19_+_50183032 | 0.04 |
ENST00000527412.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr12_-_56615693 | 0.04 |
ENST00000394013.2
ENST00000345093.4 ENST00000551711.1 ENST00000552656.1 |
RNF41
|
ring finger protein 41 |
| chr14_-_67859422 | 0.04 |
ENST00000556532.1
|
PLEK2
|
pleckstrin 2 |
| chr3_-_64253655 | 0.04 |
ENST00000498162.1
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr3_-_130745403 | 0.04 |
ENST00000504725.1
ENST00000509060.1 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr13_+_78109884 | 0.04 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr1_+_228395755 | 0.04 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr5_+_125695805 | 0.04 |
ENST00000513040.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr12_-_81763184 | 0.04 |
ENST00000548670.1
ENST00000541570.2 ENST00000553058.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chrX_+_135618258 | 0.04 |
ENST00000440515.1
ENST00000456412.1 |
VGLL1
|
vestigial like 1 (Drosophila) |
| chr19_-_55574538 | 0.03 |
ENST00000415061.3
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr13_-_114312501 | 0.03 |
ENST00000335288.4
|
ATP4B
|
ATPase, H+/K+ exchanging, beta polypeptide |
| chr10_-_104597286 | 0.03 |
ENST00000369887.3
|
CYP17A1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr18_+_54318566 | 0.03 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr17_-_33448468 | 0.03 |
ENST00000591723.1
ENST00000593039.1 ENST00000587405.1 |
RAD51L3-RFFL
RAD51D
|
Uncharacterized protein RAD51 paralog D |
| chr4_-_184241927 | 0.03 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr16_+_20817839 | 0.03 |
ENST00000348433.6
ENST00000568501.1 ENST00000566276.1 |
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr9_+_131843377 | 0.03 |
ENST00000372546.4
ENST00000406974.3 ENST00000540102.1 |
DOLPP1
|
dolichyldiphosphatase 1 |
| chr3_-_52719546 | 0.03 |
ENST00000439181.1
ENST00000449505.1 |
PBRM1
|
polybromo 1 |
| chr10_+_133747955 | 0.03 |
ENST00000455566.1
|
PPP2R2D
|
protein phosphatase 2, regulatory subunit B, delta |
| chr18_+_3449695 | 0.03 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr7_-_48068643 | 0.03 |
ENST00000453192.2
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr3_+_111260856 | 0.03 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr11_+_128634589 | 0.03 |
ENST00000281428.8
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr17_+_15635561 | 0.03 |
ENST00000584301.1
ENST00000580596.1 ENST00000464963.1 ENST00000437605.2 ENST00000579428.1 |
TBC1D26
|
TBC1 domain family, member 26 |
| chr3_+_130745688 | 0.03 |
ENST00000510769.1
ENST00000429253.2 ENST00000356918.4 ENST00000510688.1 ENST00000511262.1 ENST00000383366.4 |
NEK11
|
NIMA-related kinase 11 |
| chr1_+_19578033 | 0.03 |
ENST00000330263.4
|
MRTO4
|
mRNA turnover 4 homolog (S. cerevisiae) |
| chr2_-_24149918 | 0.03 |
ENST00000439915.1
|
ATAD2B
|
ATPase family, AAA domain containing 2B |
| chr2_+_185463093 | 0.02 |
ENST00000302277.6
|
ZNF804A
|
zinc finger protein 804A |
| chr12_-_81763127 | 0.02 |
ENST00000541017.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr14_-_80677613 | 0.02 |
ENST00000556811.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chrX_+_17755563 | 0.02 |
ENST00000380045.3
ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr3_-_52443799 | 0.02 |
ENST00000470173.1
ENST00000296288.5 |
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr1_+_44889697 | 0.02 |
ENST00000443020.2
|
RNF220
|
ring finger protein 220 |
| chr3_+_111260954 | 0.02 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr10_+_114169299 | 0.02 |
ENST00000369410.3
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr2_+_27799389 | 0.02 |
ENST00000408964.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chrX_-_48326683 | 0.02 |
ENST00000440085.1
|
SLC38A5
|
solute carrier family 38, member 5 |
| chr10_+_92631709 | 0.02 |
ENST00000413330.1
ENST00000277882.3 |
RPP30
|
ribonuclease P/MRP 30kDa subunit |
| chr17_-_28257080 | 0.02 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr10_-_5652705 | 0.02 |
ENST00000425246.1
|
RP11-336A10.5
|
RP11-336A10.5 |
| chr1_+_67773527 | 0.02 |
ENST00000541374.1
ENST00000544434.1 |
IL12RB2
|
interleukin 12 receptor, beta 2 |
| chr1_+_144151520 | 0.02 |
ENST00000369372.4
|
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr9_-_86432547 | 0.02 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr1_+_86934526 | 0.02 |
ENST00000394711.1
|
CLCA1
|
chloride channel accessory 1 |
| chrX_-_148713440 | 0.02 |
ENST00000536359.1
ENST00000316916.8 |
TMEM185A
|
transmembrane protein 185A |
| chr16_+_81528948 | 0.02 |
ENST00000539778.2
|
CMIP
|
c-Maf inducing protein |
| chr2_+_219524473 | 0.02 |
ENST00000439945.1
ENST00000431802.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr15_+_85427903 | 0.02 |
ENST00000286749.3
ENST00000394573.1 ENST00000537703.1 |
SLC28A1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chrX_-_152939252 | 0.02 |
ENST00000340888.3
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr9_-_95056010 | 0.02 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr19_+_18530146 | 0.02 |
ENST00000348495.6
ENST00000270061.7 |
SSBP4
|
single stranded DNA binding protein 4 |
| chr9_-_99540328 | 0.02 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr6_-_112194484 | 0.01 |
ENST00000518295.1
ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr17_-_50235423 | 0.01 |
ENST00000340813.6
|
CA10
|
carbonic anhydrase X |
| chr19_+_12273866 | 0.01 |
ENST00000425827.1
ENST00000439995.1 ENST00000343979.4 ENST00000398616.2 ENST00000418338.1 |
ZNF136
|
zinc finger protein 136 |
| chr6_+_151662815 | 0.01 |
ENST00000359755.5
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr18_+_54318616 | 0.01 |
ENST00000254442.3
|
WDR7
|
WD repeat domain 7 |
| chr9_-_86323118 | 0.01 |
ENST00000376395.4
|
UBQLN1
|
ubiquilin 1 |
| chr6_-_33663474 | 0.01 |
ENST00000594414.1
|
SBP1
|
SBP1; Uncharacterized protein |
| chr1_-_32264356 | 0.01 |
ENST00000452755.2
|
SPOCD1
|
SPOC domain containing 1 |
| chr3_+_130745769 | 0.01 |
ENST00000412440.2
|
NEK11
|
NIMA-related kinase 11 |
| chr8_-_101965559 | 0.01 |
ENST00000353245.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr16_+_32264040 | 0.01 |
ENST00000398664.3
|
TP53TG3D
|
TP53 target 3D |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEIS3_TGIF2LX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.3 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.5 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 1.2 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:0099541 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.0 | 0.1 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.2 | GO:0032306 | regulation of prostaglandin secretion(GO:0032306) positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.0 | GO:1903517 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.1 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.3 | GO:0042635 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 1.2 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 1.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.4 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |