Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
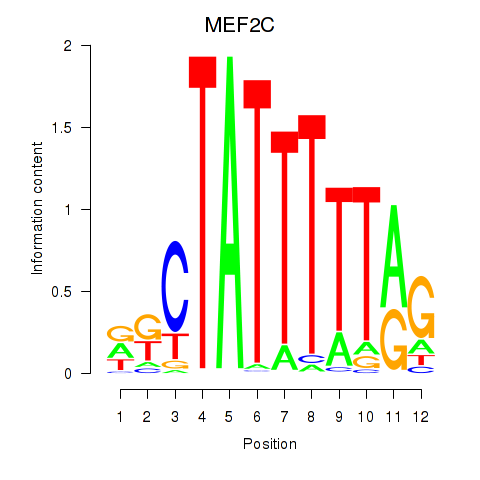
Results for MEF2C
Z-value: 0.61
Transcription factors associated with MEF2C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2C
|
ENSG00000081189.9 | myocyte enhancer factor 2C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2C | hg19_v2_chr5_-_88179302_88179319 | -0.62 | 1.9e-01 | Click! |
Activity profile of MEF2C motif
Sorted Z-values of MEF2C motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_12108410 | 0.46 |
ENST00000527997.1
|
RP13-631K18.5
|
RP13-631K18.5 |
| chr16_-_18923035 | 0.34 |
ENST00000563836.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr11_+_1860200 | 0.31 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr4_-_186696561 | 0.29 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_-_111091948 | 0.26 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr15_-_76352069 | 0.22 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr13_-_45048386 | 0.22 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr8_-_72268721 | 0.21 |
ENST00000419131.1
ENST00000388743.2 |
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr11_+_1860682 | 0.20 |
ENST00000381906.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr8_-_17533838 | 0.20 |
ENST00000400046.1
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr7_+_75931861 | 0.20 |
ENST00000248553.6
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr8_+_107282389 | 0.19 |
ENST00000577661.1
ENST00000445937.1 |
RP11-395G23.3
OXR1
|
RP11-395G23.3 oxidation resistance 1 |
| chr1_+_156095951 | 0.19 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr1_-_116383322 | 0.18 |
ENST00000429731.1
|
NHLH2
|
nescient helix loop helix 2 |
| chr6_+_151358048 | 0.18 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chrX_+_123095860 | 0.18 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr13_+_97928395 | 0.18 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr3_+_193853927 | 0.18 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr21_+_35107346 | 0.17 |
ENST00000456489.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr17_-_27467418 | 0.16 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr3_+_8543561 | 0.16 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr15_+_100106155 | 0.16 |
ENST00000557785.1
ENST00000558049.1 ENST00000449277.2 |
MEF2A
|
myocyte enhancer factor 2A |
| chr11_+_65266507 | 0.15 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr11_+_1860832 | 0.15 |
ENST00000252898.7
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr2_+_145780767 | 0.15 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr1_-_152131703 | 0.15 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr3_-_121264848 | 0.15 |
ENST00000264233.5
|
POLQ
|
polymerase (DNA directed), theta |
| chr15_+_96904487 | 0.14 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chrX_-_109683446 | 0.14 |
ENST00000372057.1
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr3_+_113616317 | 0.14 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr8_-_72268968 | 0.14 |
ENST00000388740.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr9_+_130026756 | 0.14 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr8_-_71157595 | 0.14 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr10_-_62060232 | 0.14 |
ENST00000503925.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr4_-_23891693 | 0.14 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr8_-_73793975 | 0.14 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr7_-_44105158 | 0.14 |
ENST00000297283.3
|
PGAM2
|
phosphoglycerate mutase 2 (muscle) |
| chr9_+_127054254 | 0.14 |
ENST00000422297.1
|
NEK6
|
NIMA-related kinase 6 |
| chr4_+_41362796 | 0.13 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chrX_-_119693745 | 0.13 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr14_+_62462541 | 0.13 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr15_-_52970820 | 0.13 |
ENST00000261844.7
ENST00000399202.4 ENST00000562135.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr9_+_127054217 | 0.13 |
ENST00000394199.2
ENST00000546191.1 |
NEK6
|
NIMA-related kinase 6 |
| chr12_-_57644952 | 0.12 |
ENST00000554578.1
ENST00000546246.2 ENST00000553489.1 ENST00000332782.2 |
STAC3
|
SH3 and cysteine rich domain 3 |
| chr1_+_201979743 | 0.12 |
ENST00000446188.1
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr13_+_51483814 | 0.12 |
ENST00000336617.3
ENST00000422660.1 |
RNASEH2B
|
ribonuclease H2, subunit B |
| chr14_-_23652849 | 0.12 |
ENST00000316902.7
ENST00000469263.1 ENST00000525062.1 ENST00000524758.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr8_+_1993173 | 0.11 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr2_-_148778258 | 0.11 |
ENST00000392857.5
ENST00000457954.1 ENST00000392858.1 ENST00000542387.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr7_+_120629653 | 0.11 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr1_-_204436344 | 0.11 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr8_-_72268889 | 0.11 |
ENST00000388742.4
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr5_-_142065223 | 0.11 |
ENST00000378046.1
|
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr22_+_19706958 | 0.11 |
ENST00000395109.2
|
SEPT5
|
septin 5 |
| chr9_+_100263912 | 0.11 |
ENST00000259365.4
|
TMOD1
|
tropomodulin 1 |
| chrX_-_138304939 | 0.11 |
ENST00000448673.1
|
FGF13
|
fibroblast growth factor 13 |
| chr3_-_176914238 | 0.10 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr1_+_156096336 | 0.10 |
ENST00000504687.1
ENST00000473598.2 |
LMNA
|
lamin A/C |
| chr7_+_129906660 | 0.10 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr17_+_17685422 | 0.10 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr4_+_129730779 | 0.10 |
ENST00000226319.6
|
PHF17
|
jade family PHD finger 1 |
| chr4_+_160203650 | 0.10 |
ENST00000514565.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr1_+_228395755 | 0.10 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chrX_-_131353461 | 0.10 |
ENST00000370874.1
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr5_+_67584174 | 0.10 |
ENST00000320694.8
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr12_-_57472522 | 0.10 |
ENST00000379391.3
ENST00000300128.4 |
TMEM194A
|
transmembrane protein 194A |
| chr17_-_45899126 | 0.09 |
ENST00000007414.3
ENST00000392507.3 |
OSBPL7
|
oxysterol binding protein-like 7 |
| chr3_-_176914191 | 0.09 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr3_+_36421971 | 0.09 |
ENST00000457375.2
ENST00000434649.1 |
STAC
|
SH3 and cysteine rich domain |
| chr8_+_101349823 | 0.09 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr8_+_104831472 | 0.09 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr4_-_15661474 | 0.09 |
ENST00000509314.1
ENST00000503196.1 |
FBXL5
|
F-box and leucine-rich repeat protein 5 |
| chr15_+_78830023 | 0.09 |
ENST00000599596.1
|
AC027228.1
|
HCG2005369; Uncharacterized protein |
| chr9_-_134585221 | 0.09 |
ENST00000372190.3
ENST00000427994.1 |
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr8_+_86999516 | 0.09 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr3_+_52448539 | 0.09 |
ENST00000461861.1
|
PHF7
|
PHD finger protein 7 |
| chr22_-_36031181 | 0.09 |
ENST00000594060.1
|
AL049747.1
|
AL049747.1 |
| chr14_-_61748550 | 0.09 |
ENST00000555868.1
|
TMEM30B
|
transmembrane protein 30B |
| chr2_-_235405679 | 0.09 |
ENST00000390645.2
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr19_-_893200 | 0.09 |
ENST00000269814.4
ENST00000395808.3 ENST00000312090.6 ENST00000325464.1 |
MED16
|
mediator complex subunit 16 |
| chr7_-_55583740 | 0.09 |
ENST00000453256.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr1_+_201979645 | 0.09 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr2_+_109204909 | 0.09 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr14_-_94421923 | 0.09 |
ENST00000555507.1
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chr11_+_1940925 | 0.09 |
ENST00000453458.1
ENST00000381557.2 ENST00000381589.3 ENST00000381579.3 ENST00000381563.4 ENST00000344578.4 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr10_+_32856764 | 0.08 |
ENST00000375030.2
ENST00000375028.3 |
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr15_-_70994612 | 0.08 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr7_-_124569991 | 0.08 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr5_+_173316341 | 0.08 |
ENST00000520867.1
ENST00000334035.5 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr2_+_30569506 | 0.08 |
ENST00000421976.2
|
AC109642.1
|
AC109642.1 |
| chr17_-_42200996 | 0.08 |
ENST00000587135.1
ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5
|
histone deacetylase 5 |
| chr11_+_19799327 | 0.08 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr22_+_25595817 | 0.08 |
ENST00000215855.2
ENST00000404334.1 |
CRYBB3
|
crystallin, beta B3 |
| chr4_+_160025300 | 0.08 |
ENST00000505478.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr12_+_116985896 | 0.08 |
ENST00000547114.1
|
RP11-809C9.2
|
RP11-809C9.2 |
| chr16_+_14280564 | 0.08 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr1_-_12677714 | 0.08 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr6_-_169654139 | 0.08 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chrX_+_135279179 | 0.08 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr10_+_88428370 | 0.08 |
ENST00000372066.3
ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3
|
LIM domain binding 3 |
| chr20_-_44519839 | 0.08 |
ENST00000372518.4
|
NEURL2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr4_-_25865159 | 0.08 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr18_-_19283649 | 0.08 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr15_-_93353028 | 0.07 |
ENST00000557398.2
|
FAM174B
|
family with sequence similarity 174, member B |
| chr12_+_109569155 | 0.07 |
ENST00000539864.1
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr11_-_85780853 | 0.07 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr14_+_76776957 | 0.07 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr1_+_114473350 | 0.07 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr12_-_49393092 | 0.07 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr3_-_38071122 | 0.07 |
ENST00000334661.4
|
PLCD1
|
phospholipase C, delta 1 |
| chr7_+_80267973 | 0.07 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr3_-_108248169 | 0.07 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr14_+_56046914 | 0.07 |
ENST00000413890.2
ENST00000395309.3 ENST00000554567.1 ENST00000555498.1 |
KTN1
|
kinectin 1 (kinesin receptor) |
| chr3_+_138067521 | 0.07 |
ENST00000494949.1
|
MRAS
|
muscle RAS oncogene homolog |
| chr3_+_8543533 | 0.07 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr10_+_71562180 | 0.07 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr12_+_14561422 | 0.07 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr15_+_100106244 | 0.07 |
ENST00000557942.1
|
MEF2A
|
myocyte enhancer factor 2A |
| chr4_+_41614909 | 0.07 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr15_+_78558523 | 0.07 |
ENST00000446172.2
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr8_+_1993152 | 0.07 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr20_+_58251716 | 0.07 |
ENST00000355648.4
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr14_+_39735411 | 0.07 |
ENST00000603904.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr4_+_86699834 | 0.06 |
ENST00000395183.2
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chrX_-_15683147 | 0.06 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr2_+_220299547 | 0.06 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr5_-_142065612 | 0.06 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr18_+_29027696 | 0.06 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr9_-_21995249 | 0.06 |
ENST00000494262.1
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr10_-_14880002 | 0.06 |
ENST00000465530.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr12_-_22094336 | 0.06 |
ENST00000326684.4
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr2_-_148778323 | 0.06 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr8_-_94029882 | 0.06 |
ENST00000520686.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr7_-_105332084 | 0.06 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr1_+_92632542 | 0.06 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr2_-_148779106 | 0.06 |
ENST00000416719.1
ENST00000264169.2 |
ORC4
|
origin recognition complex, subunit 4 |
| chr4_+_113970772 | 0.06 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr11_+_12766583 | 0.06 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr5_-_16509101 | 0.06 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr3_+_138067666 | 0.06 |
ENST00000475711.1
ENST00000464896.1 |
MRAS
|
muscle RAS oncogene homolog |
| chr4_+_110834033 | 0.05 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chr3_+_154797877 | 0.05 |
ENST00000462745.1
ENST00000493237.1 |
MME
|
membrane metallo-endopeptidase |
| chr14_+_56046990 | 0.05 |
ENST00000438792.2
ENST00000395314.3 ENST00000395308.1 |
KTN1
|
kinectin 1 (kinesin receptor) |
| chr15_+_100106126 | 0.05 |
ENST00000558812.1
ENST00000338042.6 |
MEF2A
|
myocyte enhancer factor 2A |
| chr3_+_4535025 | 0.05 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr2_+_232575168 | 0.05 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr17_-_79900255 | 0.05 |
ENST00000330655.3
ENST00000582198.1 |
MYADML2
PYCR1
|
myeloid-associated differentiation marker-like 2 pyrroline-5-carboxylate reductase 1 |
| chr3_-_149510553 | 0.05 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr9_+_127023704 | 0.05 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr2_-_157198860 | 0.05 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr7_-_151217166 | 0.05 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr11_+_7597639 | 0.05 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr9_+_116327326 | 0.05 |
ENST00000342620.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr7_+_152456829 | 0.05 |
ENST00000377776.3
ENST00000256001.8 ENST00000397282.2 |
ACTR3B
|
ARP3 actin-related protein 3 homolog B (yeast) |
| chr8_+_132952112 | 0.05 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr3_+_154798162 | 0.05 |
ENST00000360490.2
|
MME
|
membrane metallo-endopeptidase |
| chr15_+_80364901 | 0.05 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr15_+_40531243 | 0.05 |
ENST00000558055.1
ENST00000455577.2 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr2_+_103378472 | 0.05 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chrX_-_50386648 | 0.05 |
ENST00000460112.3
|
SHROOM4
|
shroom family member 4 |
| chr9_+_5231413 | 0.05 |
ENST00000239316.4
|
INSL4
|
insulin-like 4 (placenta) |
| chr2_-_190044480 | 0.05 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr17_-_48785216 | 0.05 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr14_+_58797974 | 0.05 |
ENST00000417477.2
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr1_-_206945830 | 0.05 |
ENST00000423557.1
|
IL10
|
interleukin 10 |
| chr9_+_97766469 | 0.05 |
ENST00000433691.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chrX_+_131157609 | 0.04 |
ENST00000496850.1
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr11_+_45918092 | 0.04 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr4_-_52883786 | 0.04 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr3_+_50316458 | 0.04 |
ENST00000316436.3
|
LSMEM2
|
leucine-rich single-pass membrane protein 2 |
| chr14_-_65409502 | 0.04 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr12_-_46766577 | 0.04 |
ENST00000256689.5
|
SLC38A2
|
solute carrier family 38, member 2 |
| chr10_-_96829246 | 0.04 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr18_+_12407895 | 0.04 |
ENST00000590956.1
ENST00000336990.4 ENST00000440960.1 ENST00000588729.1 |
SLMO1
|
slowmo homolog 1 (Drosophila) |
| chr4_-_140098339 | 0.04 |
ENST00000394235.2
|
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr9_+_97766409 | 0.04 |
ENST00000425634.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr15_+_40531621 | 0.04 |
ENST00000560346.1
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr2_+_220144168 | 0.04 |
ENST00000392087.2
ENST00000442681.1 ENST00000439026.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr7_+_152456904 | 0.04 |
ENST00000537264.1
|
ACTR3B
|
ARP3 actin-related protein 3 homolog B (yeast) |
| chr3_-_112360116 | 0.04 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr7_+_80231466 | 0.04 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr14_-_65409438 | 0.04 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr1_-_150693318 | 0.04 |
ENST00000442853.1
ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1
|
HORMA domain containing 1 |
| chrX_+_18725758 | 0.04 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chrX_+_153046456 | 0.04 |
ENST00000393786.3
ENST00000370104.1 ENST00000370108.3 ENST00000370101.3 ENST00000430541.1 ENST00000370100.1 |
SRPK3
|
SRSF protein kinase 3 |
| chr11_+_1940786 | 0.04 |
ENST00000278317.6
ENST00000381561.4 ENST00000381548.3 ENST00000360603.3 ENST00000381549.3 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr2_+_179149636 | 0.04 |
ENST00000409631.1
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr8_-_125827922 | 0.04 |
ENST00000533516.1
|
RP11-1082L8.4
|
RP11-1082L8.4 |
| chr5_+_140749803 | 0.04 |
ENST00000576222.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chr4_-_186697044 | 0.04 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_-_137475071 | 0.03 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr14_+_100204027 | 0.03 |
ENST00000554479.1
ENST00000555145.1 |
EML1
|
echinoderm microtubule associated protein like 1 |
| chr10_+_102758105 | 0.03 |
ENST00000429732.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr21_+_39628655 | 0.03 |
ENST00000398925.1
ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr1_+_230193521 | 0.03 |
ENST00000543760.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr1_-_201390846 | 0.03 |
ENST00000367312.1
ENST00000555340.2 ENST00000361379.4 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr17_-_42188598 | 0.03 |
ENST00000591714.1
|
HDAC5
|
histone deacetylase 5 |
| chr16_+_14280742 | 0.03 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr11_-_116708302 | 0.03 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr2_+_109204743 | 0.03 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2C
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:2000974 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.2 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.5 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.2 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.2 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of mitochondrial DNA metabolic process(GO:1901860) response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0035768 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.0 | GO:0060629 | regulation of homologous chromosome segregation(GO:0060629) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.0 | 0.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.0 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |