Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
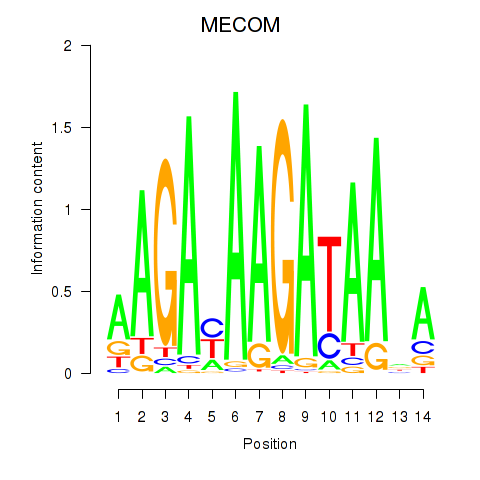
Results for MECOM
Z-value: 0.96
Transcription factors associated with MECOM
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MECOM
|
ENSG00000085276.13 | MDS1 and EVI1 complex locus |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MECOM | hg19_v2_chr3_-_169381166_169381181 | -0.63 | 1.8e-01 | Click! |
Activity profile of MECOM motif
Sorted Z-values of MECOM motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_15418467 | 0.66 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr5_+_159895275 | 0.49 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr21_+_45595354 | 0.45 |
ENST00000411956.1
|
AP001056.1
|
AP001056.1 |
| chr10_+_90484301 | 0.40 |
ENST00000404190.1
|
LIPK
|
lipase, family member K |
| chr3_-_145968857 | 0.37 |
ENST00000433593.2
ENST00000476202.1 ENST00000460885.1 |
PLSCR4
|
phospholipid scramblase 4 |
| chr12_-_15038779 | 0.36 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr11_-_33891362 | 0.35 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr3_-_145968923 | 0.34 |
ENST00000493382.1
ENST00000354952.2 ENST00000383083.2 |
PLSCR4
|
phospholipid scramblase 4 |
| chr4_+_189321881 | 0.34 |
ENST00000512839.1
ENST00000513313.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
| chr19_-_36391434 | 0.33 |
ENST00000396901.1
ENST00000585925.1 |
NFKBID
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta |
| chr6_-_132022635 | 0.31 |
ENST00000315453.2
|
OR2A4
|
olfactory receptor, family 2, subfamily A, member 4 |
| chr12_-_47219733 | 0.31 |
ENST00000547477.1
ENST00000447411.1 ENST00000266579.4 |
SLC38A4
|
solute carrier family 38, member 4 |
| chr9_+_26746951 | 0.30 |
ENST00000523363.1
|
RP11-18A15.1
|
RP11-18A15.1 |
| chr8_+_86999516 | 0.30 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr10_+_78078088 | 0.30 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chr19_-_53632912 | 0.29 |
ENST00000601215.1
|
ZNF415
|
zinc finger protein 415 |
| chr4_+_130014836 | 0.27 |
ENST00000502887.1
|
C4orf33
|
chromosome 4 open reading frame 33 |
| chr2_+_181988620 | 0.26 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr5_-_96209315 | 0.25 |
ENST00000504056.1
|
CTD-2260A17.2
|
Uncharacterized protein |
| chr4_+_117220016 | 0.25 |
ENST00000604093.1
|
MTRNR2L13
|
MT-RNR2-like 13 (pseudogene) |
| chr15_-_40401062 | 0.25 |
ENST00000354670.4
ENST00000559701.1 ENST00000557870.1 ENST00000558774.1 |
BMF
|
Bcl2 modifying factor |
| chr1_-_152539248 | 0.25 |
ENST00000368789.1
|
LCE3E
|
late cornified envelope 3E |
| chr2_+_17997763 | 0.24 |
ENST00000281047.3
|
MSGN1
|
mesogenin 1 |
| chr1_-_161193349 | 0.21 |
ENST00000469730.2
ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2
|
apolipoprotein A-II |
| chr4_-_52883786 | 0.21 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr2_+_172543919 | 0.21 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr3_-_139195350 | 0.21 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr4_+_26324474 | 0.20 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr8_-_38008783 | 0.20 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr3_-_178984759 | 0.20 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr12_-_10320190 | 0.20 |
ENST00000543993.1
ENST00000339968.6 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr13_+_102142296 | 0.20 |
ENST00000376162.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr9_-_112970436 | 0.20 |
ENST00000400613.4
|
C9orf152
|
chromosome 9 open reading frame 152 |
| chr14_-_82089405 | 0.19 |
ENST00000554211.1
|
RP11-799P8.1
|
RP11-799P8.1 |
| chr12_+_28605426 | 0.19 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr6_+_49431073 | 0.19 |
ENST00000335783.3
|
CENPQ
|
centromere protein Q |
| chr4_-_122148620 | 0.19 |
ENST00000509841.1
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr8_+_97773202 | 0.18 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr3_+_177534653 | 0.18 |
ENST00000436078.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr2_+_172543967 | 0.18 |
ENST00000534253.2
ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr19_-_35981358 | 0.17 |
ENST00000484218.2
ENST00000338897.3 |
KRTDAP
|
keratinocyte differentiation-associated protein |
| chr8_-_6420565 | 0.17 |
ENST00000338312.6
|
ANGPT2
|
angiopoietin 2 |
| chr5_+_67485704 | 0.16 |
ENST00000520762.1
|
RP11-404L6.2
|
Uncharacterized protein |
| chr1_-_98386543 | 0.16 |
ENST00000423006.2
ENST00000370192.3 ENST00000306031.5 |
DPYD
|
dihydropyrimidine dehydrogenase |
| chr17_+_70036164 | 0.16 |
ENST00000602013.1
|
AC007461.1
|
Uncharacterized protein |
| chr4_+_26323764 | 0.16 |
ENST00000514730.1
ENST00000507574.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr15_+_39311432 | 0.16 |
ENST00000560709.1
|
RP11-62C7.2
|
RP11-62C7.2 |
| chr3_-_119379427 | 0.15 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr1_-_68962744 | 0.15 |
ENST00000525124.1
|
DEPDC1
|
DEP domain containing 1 |
| chr6_-_75912508 | 0.15 |
ENST00000416123.2
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr12_+_113344811 | 0.15 |
ENST00000551241.1
ENST00000553185.1 ENST00000550689.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr10_-_14596140 | 0.14 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr15_+_40532837 | 0.14 |
ENST00000560806.1
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr1_+_207038699 | 0.14 |
ENST00000367098.1
|
IL20
|
interleukin 20 |
| chrX_-_149999722 | 0.14 |
ENST00000418547.1
|
CD99L2
|
CD99 molecule-like 2 |
| chr4_+_96012585 | 0.13 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr19_+_23257988 | 0.13 |
ENST00000594318.1
ENST00000600223.1 ENST00000593635.1 |
CTD-2291D10.4
ZNF730
|
CTD-2291D10.4 zinc finger protein 730 |
| chr10_-_5652705 | 0.13 |
ENST00000425246.1
|
RP11-336A10.5
|
RP11-336A10.5 |
| chr12_-_8815299 | 0.13 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr5_+_171621176 | 0.13 |
ENST00000398186.4
|
EFCAB9
|
EF-hand calcium binding domain 9 |
| chr1_+_171060018 | 0.12 |
ENST00000367755.4
ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3
|
flavin containing monooxygenase 3 |
| chr9_+_127023704 | 0.12 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chrX_-_43741594 | 0.12 |
ENST00000536181.1
ENST00000378069.4 |
MAOB
|
monoamine oxidase B |
| chr13_+_31309645 | 0.12 |
ENST00000380490.3
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein |
| chr2_-_157189180 | 0.12 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr1_-_45452240 | 0.12 |
ENST00000372183.3
ENST00000372182.4 ENST00000360403.2 |
EIF2B3
|
eukaryotic translation initiation factor 2B, subunit 3 gamma, 58kDa |
| chr12_-_76879852 | 0.12 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr15_-_38852251 | 0.12 |
ENST00000558432.1
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr12_-_12715266 | 0.12 |
ENST00000228862.2
|
DUSP16
|
dual specificity phosphatase 16 |
| chr12_+_27863706 | 0.12 |
ENST00000081029.3
ENST00000538315.1 ENST00000542791.1 |
MRPS35
|
mitochondrial ribosomal protein S35 |
| chrX_-_19765692 | 0.12 |
ENST00000432234.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr15_-_83837983 | 0.12 |
ENST00000562702.1
|
HDGFRP3
|
Hepatoma-derived growth factor-related protein 3 |
| chr3_-_145968900 | 0.12 |
ENST00000460350.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr2_+_172544011 | 0.11 |
ENST00000508530.1
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr16_+_9449445 | 0.11 |
ENST00000564305.1
|
RP11-243A14.1
|
RP11-243A14.1 |
| chr7_-_15601595 | 0.11 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr13_-_113242439 | 0.11 |
ENST00000375669.3
ENST00000261965.3 |
TUBGCP3
|
tubulin, gamma complex associated protein 3 |
| chr18_-_24722995 | 0.11 |
ENST00000581714.1
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr3_-_161089289 | 0.10 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr7_-_8302207 | 0.10 |
ENST00000407906.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr15_-_55562451 | 0.10 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr10_+_116524547 | 0.10 |
ENST00000436932.1
|
RP11-106M7.1
|
RP11-106M7.1 |
| chr22_+_29138013 | 0.10 |
ENST00000216027.3
ENST00000398941.2 |
HSCB
|
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr15_-_48470544 | 0.10 |
ENST00000267836.6
|
MYEF2
|
myelin expression factor 2 |
| chr17_+_38121772 | 0.10 |
ENST00000577447.1
|
GSDMA
|
gasdermin A |
| chr6_-_52860171 | 0.09 |
ENST00000370963.4
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chrX_+_15525426 | 0.09 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr6_-_52859968 | 0.09 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr5_+_125695805 | 0.09 |
ENST00000513040.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr1_-_52520828 | 0.09 |
ENST00000610127.1
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr5_-_127674883 | 0.09 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr2_+_190541153 | 0.09 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr8_+_105602961 | 0.09 |
ENST00000521923.1
|
RP11-127H5.1
|
Uncharacterized protein |
| chr5_+_56471592 | 0.09 |
ENST00000511209.1
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chrX_+_155227246 | 0.09 |
ENST00000244174.5
ENST00000424344.3 |
IL9R
|
interleukin 9 receptor |
| chr5_+_121465207 | 0.09 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr16_-_80673633 | 0.08 |
ENST00000561616.1
|
CDYL2
|
chromodomain protein, Y-like 2 |
| chr1_-_110155671 | 0.08 |
ENST00000351050.3
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr7_-_100844193 | 0.08 |
ENST00000440203.2
ENST00000379423.3 ENST00000223114.4 |
MOGAT3
|
monoacylglycerol O-acyltransferase 3 |
| chr11_+_58695096 | 0.08 |
ENST00000525608.1
ENST00000526351.1 |
GLYATL1
|
glycine-N-acyltransferase-like 1 |
| chr5_-_64920115 | 0.08 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr12_-_86650077 | 0.08 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr2_+_242289502 | 0.08 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr3_+_99986036 | 0.08 |
ENST00000471098.1
|
TBC1D23
|
TBC1 domain family, member 23 |
| chr15_+_100348193 | 0.08 |
ENST00000558188.1
|
CTD-2054N24.2
|
Uncharacterized protein |
| chr12_-_112443830 | 0.08 |
ENST00000550037.1
ENST00000549425.1 |
TMEM116
|
transmembrane protein 116 |
| chr7_-_8302164 | 0.08 |
ENST00000447326.1
ENST00000406470.2 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr1_+_47901689 | 0.08 |
ENST00000334793.5
|
FOXD2
|
forkhead box D2 |
| chr1_+_248031277 | 0.07 |
ENST00000537741.1
|
OR2W3
|
olfactory receptor, family 2, subfamily W, member 3 |
| chr5_-_133340682 | 0.07 |
ENST00000265333.3
|
VDAC1
|
voltage-dependent anion channel 1 |
| chr15_-_48470558 | 0.07 |
ENST00000324324.7
|
MYEF2
|
myelin expression factor 2 |
| chr2_+_27805971 | 0.07 |
ENST00000413371.2
|
ZNF512
|
zinc finger protein 512 |
| chr6_+_27215494 | 0.07 |
ENST00000230582.3
|
PRSS16
|
protease, serine, 16 (thymus) |
| chr3_+_152552685 | 0.07 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr7_+_44240520 | 0.07 |
ENST00000496112.1
ENST00000223369.2 |
YKT6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr2_-_1629176 | 0.07 |
ENST00000366424.2
|
AC144450.2
|
AC144450.2 |
| chr12_-_39734783 | 0.07 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr4_-_100065419 | 0.07 |
ENST00000504125.1
ENST00000505590.1 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr16_+_28943260 | 0.07 |
ENST00000538922.1
ENST00000324662.3 ENST00000567541.1 |
CD19
|
CD19 molecule |
| chr2_-_237416181 | 0.06 |
ENST00000409907.3
|
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr3_+_99357319 | 0.06 |
ENST00000452013.1
ENST00000261037.3 ENST00000273342.4 |
COL8A1
|
collagen, type VIII, alpha 1 |
| chr5_-_138862326 | 0.06 |
ENST00000330794.4
|
TMEM173
|
transmembrane protein 173 |
| chr3_-_178103144 | 0.06 |
ENST00000417383.1
ENST00000418585.1 ENST00000411727.1 ENST00000439810.1 |
RP11-33A14.1
|
RP11-33A14.1 |
| chr5_+_20616500 | 0.06 |
ENST00000512688.1
|
RP11-774D14.1
|
RP11-774D14.1 |
| chr6_-_154831779 | 0.06 |
ENST00000607772.1
|
CNKSR3
|
CNKSR family member 3 |
| chr17_+_22022437 | 0.06 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr22_-_31688431 | 0.06 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr9_-_13175823 | 0.06 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr2_+_108145913 | 0.06 |
ENST00000443205.1
|
AC096669.3
|
AC096669.3 |
| chrX_+_129473916 | 0.06 |
ENST00000545805.1
ENST00000543953.1 ENST00000218197.5 |
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr20_-_45984401 | 0.06 |
ENST00000311275.7
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr1_+_218683438 | 0.06 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr11_+_94300474 | 0.06 |
ENST00000299001.6
|
PIWIL4
|
piwi-like RNA-mediated gene silencing 4 |
| chr5_+_96211643 | 0.06 |
ENST00000437043.3
ENST00000510373.1 |
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr2_-_237416071 | 0.06 |
ENST00000309507.5
ENST00000431676.2 |
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr1_+_160160283 | 0.05 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr15_-_30261066 | 0.05 |
ENST00000558447.1
|
TJP1
|
tight junction protein 1 |
| chr14_-_25479811 | 0.05 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr3_-_109035342 | 0.05 |
ENST00000478945.1
|
DPPA2
|
developmental pluripotency associated 2 |
| chr12_+_53836339 | 0.05 |
ENST00000549135.1
|
PRR13
|
proline rich 13 |
| chr3_+_119013185 | 0.05 |
ENST00000264245.4
|
ARHGAP31
|
Rho GTPase activating protein 31 |
| chr19_-_54106751 | 0.05 |
ENST00000600193.1
|
CTB-167G5.5
|
Uncharacterized protein |
| chr1_+_160160346 | 0.05 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr9_+_109213096 | 0.05 |
ENST00000425709.1
|
RP11-308N19.1
|
RP11-308N19.1 |
| chr5_+_167956121 | 0.05 |
ENST00000338333.4
|
FBLL1
|
fibrillarin-like 1 |
| chr14_-_69261310 | 0.05 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr15_-_30686052 | 0.05 |
ENST00000562729.1
|
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr3_+_4344988 | 0.05 |
ENST00000358065.4
|
SETMAR
|
SET domain and mariner transposase fusion gene |
| chr1_-_152009460 | 0.05 |
ENST00000271638.2
|
S100A11
|
S100 calcium binding protein A11 |
| chr1_+_154955797 | 0.05 |
ENST00000368433.1
ENST00000315144.10 ENST00000368432.1 ENST00000368431.3 ENST00000292180.3 |
FLAD1
|
flavin adenine dinucleotide synthetase 1 |
| chr10_-_18940501 | 0.05 |
ENST00000377304.4
|
NSUN6
|
NOP2/Sun domain family, member 6 |
| chr17_-_29648761 | 0.05 |
ENST00000247270.3
ENST00000462804.2 |
EVI2A
|
ecotropic viral integration site 2A |
| chr3_+_39448180 | 0.05 |
ENST00000301821.6
ENST00000458478.1 ENST00000443003.1 |
RPSA
|
ribosomal protein SA |
| chr20_-_45985414 | 0.05 |
ENST00000461685.1
ENST00000372023.3 ENST00000540497.1 ENST00000435836.1 ENST00000471951.2 ENST00000352431.2 ENST00000396281.4 ENST00000355972.4 ENST00000360911.3 |
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr8_+_72740402 | 0.05 |
ENST00000521467.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr1_-_157069590 | 0.05 |
ENST00000454449.2
|
ETV3L
|
ets variant 3-like |
| chr11_+_47586982 | 0.05 |
ENST00000426530.2
ENST00000534775.1 |
PTPMT1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr2_+_27805880 | 0.05 |
ENST00000379717.1
ENST00000355467.4 ENST00000556601.1 ENST00000416005.2 |
ZNF512
|
zinc finger protein 512 |
| chr15_+_78441663 | 0.05 |
ENST00000299518.2
ENST00000558554.1 ENST00000557826.1 ENST00000561279.1 ENST00000559186.1 ENST00000560770.1 ENST00000559881.1 ENST00000559205.1 ENST00000441490.2 |
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr1_-_85156417 | 0.05 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr19_-_53632925 | 0.05 |
ENST00000595174.1
|
ZNF415
|
zinc finger protein 415 |
| chr12_+_113344582 | 0.05 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr16_+_71879861 | 0.05 |
ENST00000427980.2
ENST00000568581.1 |
ATXN1L
IST1
|
ataxin 1-like increased sodium tolerance 1 homolog (yeast) |
| chr19_-_48894762 | 0.05 |
ENST00000600980.1
ENST00000330720.2 |
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr2_-_56150910 | 0.04 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr4_-_40859132 | 0.04 |
ENST00000543538.1
ENST00000502841.1 ENST00000504305.1 ENST00000513516.1 ENST00000510670.1 |
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr8_+_22857048 | 0.04 |
ENST00000251822.6
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr20_-_45985172 | 0.04 |
ENST00000536340.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr11_+_12115543 | 0.04 |
ENST00000537344.1
ENST00000532179.1 ENST00000526065.1 |
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr14_+_24584056 | 0.04 |
ENST00000561001.1
|
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr13_+_111855399 | 0.04 |
ENST00000426768.2
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr11_+_65266507 | 0.04 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr22_+_30279144 | 0.04 |
ENST00000401950.2
ENST00000333027.3 ENST00000445401.1 ENST00000323630.5 ENST00000351488.3 |
MTMR3
|
myotubularin related protein 3 |
| chr12_+_12202774 | 0.04 |
ENST00000589718.1
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr11_+_58695174 | 0.04 |
ENST00000317391.4
|
GLYATL1
|
glycine-N-acyltransferase-like 1 |
| chr5_+_92919043 | 0.04 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr1_-_208084729 | 0.04 |
ENST00000310833.7
ENST00000356522.4 |
CD34
|
CD34 molecule |
| chr5_-_115910091 | 0.04 |
ENST00000257414.8
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr7_-_6007070 | 0.04 |
ENST00000337579.3
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr11_+_47587129 | 0.04 |
ENST00000326656.8
ENST00000326674.9 |
PTPMT1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr21_-_28338732 | 0.04 |
ENST00000284987.5
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
| chr8_+_134203303 | 0.04 |
ENST00000519433.1
ENST00000517423.1 ENST00000377863.2 ENST00000220856.6 |
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chrX_-_19905577 | 0.04 |
ENST00000379697.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr3_+_108541545 | 0.04 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr15_-_55562479 | 0.04 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_+_21207503 | 0.04 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr3_-_193272741 | 0.04 |
ENST00000392443.3
|
ATP13A4
|
ATPase type 13A4 |
| chr15_-_102285007 | 0.04 |
ENST00000560292.2
|
RP11-89K11.1
|
Uncharacterized protein |
| chr5_+_140165876 | 0.04 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr2_-_38056538 | 0.04 |
ENST00000413792.1
|
LINC00211
|
long intergenic non-protein coding RNA 211 |
| chr3_-_111314230 | 0.04 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr1_+_172422026 | 0.04 |
ENST00000367725.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr2_-_224467093 | 0.03 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chrX_+_86772787 | 0.03 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chr1_+_161035655 | 0.03 |
ENST00000600454.1
|
AL591806.1
|
Uncharacterized protein |
| chr8_+_134203273 | 0.03 |
ENST00000250160.6
|
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr4_-_100065389 | 0.03 |
ENST00000512499.1
|
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr14_-_38028689 | 0.03 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr8_-_6420930 | 0.03 |
ENST00000325203.5
|
ANGPT2
|
angiopoietin 2 |
| chr8_-_17942432 | 0.03 |
ENST00000381733.4
ENST00000314146.10 |
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MECOM
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:2000909 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of cholesterol import(GO:0060620) regulation of intestinal lipid absorption(GO:1904729) regulation of sterol import(GO:2000909) |
| 0.1 | 0.2 | GO:0018963 | insecticide metabolic process(GO:0017143) phthalate metabolic process(GO:0018963) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.4 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) |
| 0.0 | 0.5 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.8 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.1 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.2 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) regulation of penile erection(GO:0060405) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0072387 | flavin-containing compound biosynthetic process(GO:0042727) flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.0 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.0 | GO:0072011 | glomerular endothelium development(GO:0072011) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.0 | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.0 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.0 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.1 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.1 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0047946 | glutamine N-acyltransferase activity(GO:0047946) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.4 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.0 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0005046 | KDEL sequence binding(GO:0005046) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |