Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
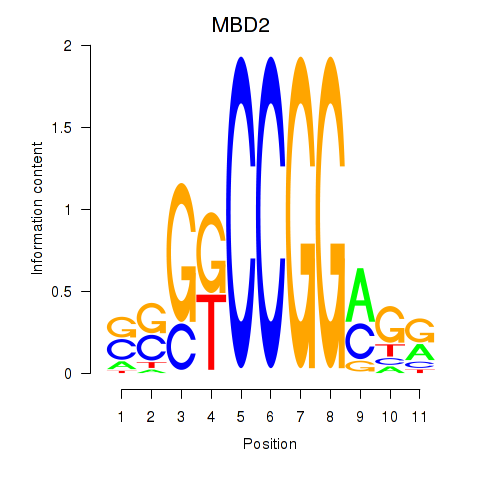
Results for MBD2
Z-value: 1.16
Transcription factors associated with MBD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MBD2
|
ENSG00000134046.7 | methyl-CpG binding domain protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MBD2 | hg19_v2_chr18_-_51751132_51751158 | -0.91 | 1.2e-02 | Click! |
Activity profile of MBD2 motif
Sorted Z-values of MBD2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_74212073 | 1.90 |
ENST00000441217.1
|
AC073046.25
|
AC073046.25 |
| chr19_-_14201507 | 1.04 |
ENST00000533683.2
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr19_-_14201776 | 0.69 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr8_+_61592073 | 0.53 |
ENST00000526846.1
|
CHD7
|
chromodomain helicase DNA binding protein 7 |
| chr16_+_2022036 | 0.50 |
ENST00000568546.1
|
TBL3
|
transducin (beta)-like 3 |
| chr1_-_22469459 | 0.49 |
ENST00000290167.6
|
WNT4
|
wingless-type MMTV integration site family, member 4 |
| chr18_-_72264805 | 0.46 |
ENST00000577806.1
|
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chr22_-_19419205 | 0.46 |
ENST00000340170.4
ENST00000263208.5 |
HIRA
|
histone cell cycle regulator |
| chr10_+_104180580 | 0.46 |
ENST00000425536.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr16_+_2059872 | 0.45 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr6_+_139349903 | 0.45 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like |
| chr10_+_129845670 | 0.45 |
ENST00000467366.1
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr12_-_58159361 | 0.44 |
ENST00000546567.1
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr11_+_64685026 | 0.43 |
ENST00000526559.1
|
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta |
| chr1_-_156698591 | 0.39 |
ENST00000368219.1
|
ISG20L2
|
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr10_-_17659234 | 0.38 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr1_+_43148059 | 0.37 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr17_-_4889508 | 0.35 |
ENST00000574606.2
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr8_-_145688231 | 0.35 |
ENST00000530374.1
|
CYHR1
|
cysteine/histidine-rich 1 |
| chr16_-_30905584 | 0.35 |
ENST00000380317.4
|
BCL7C
|
B-cell CLL/lymphoma 7C |
| chr6_-_43596899 | 0.35 |
ENST00000307126.5
ENST00000452781.1 |
GTPBP2
|
GTP binding protein 2 |
| chr12_+_122064673 | 0.35 |
ENST00000537188.1
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr8_-_57123815 | 0.34 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr6_+_2245982 | 0.34 |
ENST00000530346.1
ENST00000524770.1 ENST00000532124.1 ENST00000531092.1 ENST00000456943.2 ENST00000529893.1 |
GMDS-AS1
|
GMDS antisense RNA 1 (head to head) |
| chr1_-_156698181 | 0.34 |
ENST00000313146.6
|
ISG20L2
|
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr2_+_181845532 | 0.34 |
ENST00000602475.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr3_-_39195037 | 0.33 |
ENST00000273153.5
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1 |
| chr19_+_36545833 | 0.32 |
ENST00000401500.2
ENST00000270301.7 ENST00000427823.2 |
WDR62
|
WD repeat domain 62 |
| chr19_+_56152361 | 0.32 |
ENST00000585995.1
ENST00000592996.1 |
ZNF581
CCDC106
|
zinc finger protein 581 coiled-coil domain containing 106 |
| chr3_+_112930946 | 0.30 |
ENST00000462425.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr6_+_17281573 | 0.30 |
ENST00000379052.5
|
RBM24
|
RNA binding motif protein 24 |
| chr11_+_64851729 | 0.30 |
ENST00000526791.1
ENST00000526945.1 |
ZFPL1
|
zinc finger protein-like 1 |
| chr5_+_14143728 | 0.29 |
ENST00000344204.4
ENST00000537187.1 |
TRIO
|
trio Rho guanine nucleotide exchange factor |
| chr17_-_42276574 | 0.29 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr9_+_103189405 | 0.29 |
ENST00000395067.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr12_-_49453557 | 0.29 |
ENST00000547610.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr12_-_117319236 | 0.29 |
ENST00000257572.5
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr14_+_103851712 | 0.28 |
ENST00000440884.3
ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3
|
MAP/microtubule affinity-regulating kinase 3 |
| chr11_+_76092353 | 0.28 |
ENST00000530460.1
ENST00000321844.4 |
RP11-111M22.2
|
Homo sapiens putative uncharacterized protein FLJ37770-like (LOC100506127), mRNA. |
| chr17_+_27920486 | 0.28 |
ENST00000394859.3
|
ANKRD13B
|
ankyrin repeat domain 13B |
| chr2_-_46769694 | 0.28 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr11_-_75236867 | 0.28 |
ENST00000376282.3
ENST00000336898.3 |
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr17_+_54671047 | 0.28 |
ENST00000332822.4
|
NOG
|
noggin |
| chr10_+_104404218 | 0.27 |
ENST00000302424.7
|
TRIM8
|
tripartite motif containing 8 |
| chr19_+_36606354 | 0.27 |
ENST00000589996.1
ENST00000591296.1 |
TBCB
|
tubulin folding cofactor B |
| chr5_-_176778523 | 0.27 |
ENST00000513877.1
ENST00000515209.1 ENST00000514458.1 ENST00000502560.1 |
LMAN2
|
lectin, mannose-binding 2 |
| chr8_-_67525524 | 0.27 |
ENST00000517885.1
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr20_-_48532019 | 0.27 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr1_+_43148625 | 0.26 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr12_-_57400227 | 0.26 |
ENST00000300101.2
|
ZBTB39
|
zinc finger and BTB domain containing 39 |
| chr16_-_2059797 | 0.26 |
ENST00000563630.1
|
ZNF598
|
zinc finger protein 598 |
| chr1_+_11539204 | 0.25 |
ENST00000294484.6
ENST00000389575.3 |
PTCHD2
|
patched domain containing 2 |
| chr16_-_2059748 | 0.25 |
ENST00000562103.1
ENST00000431526.1 |
ZNF598
|
zinc finger protein 598 |
| chr19_-_46476791 | 0.25 |
ENST00000263257.5
|
NOVA2
|
neuro-oncological ventral antigen 2 |
| chr20_+_3767547 | 0.25 |
ENST00000344256.6
ENST00000379598.5 |
CDC25B
|
cell division cycle 25B |
| chr11_-_89224638 | 0.24 |
ENST00000535633.1
ENST00000263317.4 |
NOX4
|
NADPH oxidase 4 |
| chr8_-_141645645 | 0.24 |
ENST00000519980.1
ENST00000220592.5 |
AGO2
|
argonaute RISC catalytic component 2 |
| chr11_-_75141206 | 0.24 |
ENST00000376292.4
|
KLHL35
|
kelch-like family member 35 |
| chr22_-_39096661 | 0.24 |
ENST00000216039.5
|
JOSD1
|
Josephin domain containing 1 |
| chr17_+_43299241 | 0.23 |
ENST00000328118.3
|
FMNL1
|
formin-like 1 |
| chr11_+_64851666 | 0.23 |
ENST00000525509.1
ENST00000294258.3 ENST00000526334.1 |
ZFPL1
|
zinc finger protein-like 1 |
| chr14_+_53019822 | 0.23 |
ENST00000321662.6
|
GPR137C
|
G protein-coupled receptor 137C |
| chr1_-_40237020 | 0.22 |
ENST00000327582.5
|
OXCT2
|
3-oxoacid CoA transferase 2 |
| chr9_+_103189458 | 0.22 |
ENST00000398977.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr14_-_103523745 | 0.22 |
ENST00000361246.2
|
CDC42BPB
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr22_+_29279552 | 0.22 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr10_-_44070016 | 0.22 |
ENST00000374446.2
ENST00000426961.1 ENST00000535642.1 |
ZNF239
|
zinc finger protein 239 |
| chr19_-_36606181 | 0.21 |
ENST00000221859.4
|
POLR2I
|
polymerase (RNA) II (DNA directed) polypeptide I, 14.5kDa |
| chr19_+_35491174 | 0.21 |
ENST00000317991.5
ENST00000504615.2 |
GRAMD1A
|
GRAM domain containing 1A |
| chr22_+_41487711 | 0.21 |
ENST00000263253.7
|
EP300
|
E1A binding protein p300 |
| chr10_+_75545391 | 0.21 |
ENST00000604524.1
ENST00000605216.1 ENST00000398706.2 |
ZSWIM8
|
zinc finger, SWIM-type containing 8 |
| chrX_+_12156582 | 0.21 |
ENST00000380682.1
|
FRMPD4
|
FERM and PDZ domain containing 4 |
| chr12_-_29936731 | 0.21 |
ENST00000552618.1
ENST00000539277.1 ENST00000551659.1 |
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr20_-_56284816 | 0.21 |
ENST00000395819.3
ENST00000341744.3 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr12_+_7037461 | 0.21 |
ENST00000396684.2
|
ATN1
|
atrophin 1 |
| chr6_+_1389989 | 0.21 |
ENST00000259806.1
|
FOXF2
|
forkhead box F2 |
| chr14_+_23564700 | 0.21 |
ENST00000554203.1
|
C14orf119
|
chromosome 14 open reading frame 119 |
| chr6_+_125474992 | 0.21 |
ENST00000528193.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr2_+_220306238 | 0.21 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr17_+_6544356 | 0.20 |
ENST00000574838.1
|
TXNDC17
|
thioredoxin domain containing 17 |
| chr1_+_3689325 | 0.20 |
ENST00000444870.2
ENST00000452264.1 |
SMIM1
|
small integral membrane protein 1 (Vel blood group) |
| chr1_-_32801825 | 0.20 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr6_-_91296737 | 0.20 |
ENST00000369332.3
ENST00000369329.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr19_-_46148820 | 0.20 |
ENST00000587152.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr22_+_19419425 | 0.20 |
ENST00000333130.3
|
MRPL40
|
mitochondrial ribosomal protein L40 |
| chr19_-_10341948 | 0.20 |
ENST00000590320.1
ENST00000592342.1 ENST00000588952.1 |
S1PR2
DNMT1
|
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr10_+_115803650 | 0.20 |
ENST00000369295.2
|
ADRB1
|
adrenoceptor beta 1 |
| chr12_-_93835665 | 0.20 |
ENST00000552442.1
ENST00000550657.1 |
UBE2N
|
ubiquitin-conjugating enzyme E2N |
| chr20_-_1447467 | 0.19 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr19_+_56152262 | 0.19 |
ENST00000325333.5
ENST00000590190.1 |
ZNF580
|
zinc finger protein 580 |
| chr19_-_4124079 | 0.19 |
ENST00000394867.4
ENST00000262948.5 |
MAP2K2
|
mitogen-activated protein kinase kinase 2 |
| chr20_-_39317868 | 0.19 |
ENST00000373313.2
|
MAFB
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr19_+_36545781 | 0.19 |
ENST00000388999.3
|
WDR62
|
WD repeat domain 62 |
| chr10_+_75545329 | 0.19 |
ENST00000604729.1
ENST00000603114.1 |
ZSWIM8
|
zinc finger, SWIM-type containing 8 |
| chr17_-_29151686 | 0.19 |
ENST00000544695.1
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr3_+_184097836 | 0.19 |
ENST00000204604.1
ENST00000310236.3 |
CHRD
|
chordin |
| chr14_-_77787198 | 0.19 |
ENST00000261534.4
|
POMT2
|
protein-O-mannosyltransferase 2 |
| chr16_+_57769635 | 0.19 |
ENST00000379661.3
ENST00000562592.1 ENST00000566726.1 |
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr1_-_27693349 | 0.19 |
ENST00000374040.3
ENST00000357582.2 ENST00000493901.1 |
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr9_+_100745615 | 0.19 |
ENST00000339399.4
|
ANP32B
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr1_+_43855560 | 0.18 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr17_-_42277203 | 0.18 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr1_+_226411319 | 0.18 |
ENST00000542034.1
ENST00000366810.5 |
MIXL1
|
Mix paired-like homeobox |
| chr2_+_5832799 | 0.18 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chr20_-_30310797 | 0.18 |
ENST00000422920.1
|
BCL2L1
|
BCL2-like 1 |
| chr6_+_42981922 | 0.18 |
ENST00000326974.4
ENST00000244670.8 |
KLHDC3
|
kelch domain containing 3 |
| chr19_+_18496957 | 0.18 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr15_-_80215984 | 0.18 |
ENST00000485386.1
ENST00000479961.1 |
ST20
ST20-MTHFS
|
suppressor of tumorigenicity 20 ST20-MTHFS readthrough |
| chr3_+_14989076 | 0.18 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr2_-_219537134 | 0.18 |
ENST00000295704.2
|
RNF25
|
ring finger protein 25 |
| chr5_+_76506706 | 0.18 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr12_+_122516626 | 0.18 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chr9_+_132597722 | 0.18 |
ENST00000372429.3
ENST00000315480.4 ENST00000358355.1 |
USP20
|
ubiquitin specific peptidase 20 |
| chr8_+_37654424 | 0.18 |
ENST00000315215.7
|
GPR124
|
G protein-coupled receptor 124 |
| chr19_-_49622348 | 0.18 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr1_+_179851893 | 0.18 |
ENST00000531630.2
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chr16_-_28074822 | 0.18 |
ENST00000395724.3
ENST00000380898.2 ENST00000447459.2 |
GSG1L
|
GSG1-like |
| chr17_+_36861735 | 0.18 |
ENST00000378137.5
ENST00000325718.7 |
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr19_+_36208877 | 0.18 |
ENST00000420124.1
ENST00000222270.7 ENST00000341701.1 |
KMT2B
|
Histone-lysine N-methyltransferase 2B |
| chr14_+_103243813 | 0.18 |
ENST00000560371.1
ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3
|
TNF receptor-associated factor 3 |
| chr17_-_80231557 | 0.17 |
ENST00000392334.2
ENST00000314028.6 |
CSNK1D
|
casein kinase 1, delta |
| chr3_+_133292851 | 0.17 |
ENST00000503932.1
|
CDV3
|
CDV3 homolog (mouse) |
| chr20_-_47804894 | 0.17 |
ENST00000371828.3
ENST00000371856.2 ENST00000360426.4 ENST00000347458.5 ENST00000340954.7 ENST00000371802.1 ENST00000371792.1 ENST00000437404.2 |
STAU1
|
staufen double-stranded RNA binding protein 1 |
| chr5_-_176900610 | 0.17 |
ENST00000477391.2
ENST00000393565.1 ENST00000309007.5 |
DBN1
|
drebrin 1 |
| chr11_+_65383227 | 0.17 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr11_-_67141090 | 0.17 |
ENST00000312438.7
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr5_+_167718604 | 0.17 |
ENST00000265293.4
|
WWC1
|
WW and C2 domain containing 1 |
| chr4_-_170192000 | 0.17 |
ENST00000502315.1
|
SH3RF1
|
SH3 domain containing ring finger 1 |
| chr17_-_41623259 | 0.17 |
ENST00000538265.1
ENST00000591713.1 |
ETV4
|
ets variant 4 |
| chr11_-_2160611 | 0.16 |
ENST00000416167.2
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr11_+_111473108 | 0.16 |
ENST00000304987.3
|
SIK2
|
salt-inducible kinase 2 |
| chr22_-_42739533 | 0.16 |
ENST00000515426.1
|
TCF20
|
transcription factor 20 (AR1) |
| chr4_+_6202448 | 0.16 |
ENST00000508601.1
|
RP11-586D19.1
|
RP11-586D19.1 |
| chr2_-_208031542 | 0.16 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chrX_+_68725084 | 0.16 |
ENST00000252338.4
|
FAM155B
|
family with sequence similarity 155, member B |
| chr20_-_1447547 | 0.16 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr3_+_184097905 | 0.16 |
ENST00000450923.1
|
CHRD
|
chordin |
| chr20_+_34894247 | 0.16 |
ENST00000373913.3
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4 |
| chr19_+_35491330 | 0.16 |
ENST00000411896.2
ENST00000424536.2 |
GRAMD1A
|
GRAM domain containing 1A |
| chr16_+_2732476 | 0.16 |
ENST00000301738.4
ENST00000564195.1 |
KCTD5
|
potassium channel tetramerization domain containing 5 |
| chr17_-_41623009 | 0.16 |
ENST00000393664.2
|
ETV4
|
ets variant 4 |
| chr12_+_50479101 | 0.16 |
ENST00000551966.1
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr16_+_67062996 | 0.16 |
ENST00000561924.2
|
CBFB
|
core-binding factor, beta subunit |
| chr3_+_71803201 | 0.16 |
ENST00000304411.2
|
GPR27
|
G protein-coupled receptor 27 |
| chr5_+_156569944 | 0.16 |
ENST00000521769.1
|
ITK
|
IL2-inducible T-cell kinase |
| chr15_+_90544532 | 0.16 |
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710 |
| chr6_+_44238203 | 0.16 |
ENST00000451188.2
|
TMEM151B
|
transmembrane protein 151B |
| chr8_-_93115445 | 0.16 |
ENST00000523629.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr19_-_11373128 | 0.16 |
ENST00000294618.7
|
DOCK6
|
dedicator of cytokinesis 6 |
| chr19_+_38879061 | 0.16 |
ENST00000587013.1
|
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr19_+_49622646 | 0.15 |
ENST00000334186.4
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr16_+_89787393 | 0.15 |
ENST00000289816.5
ENST00000568064.1 |
ZNF276
|
zinc finger protein 276 |
| chr1_+_156698708 | 0.15 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr20_-_48532046 | 0.15 |
ENST00000543716.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr17_-_48227877 | 0.15 |
ENST00000316878.6
|
PPP1R9B
|
protein phosphatase 1, regulatory subunit 9B |
| chr17_+_43299156 | 0.15 |
ENST00000331495.3
|
FMNL1
|
formin-like 1 |
| chr7_+_129251531 | 0.15 |
ENST00000393232.1
ENST00000353868.4 ENST00000539636.1 ENST00000454688.1 ENST00000223190.4 ENST00000311967.2 |
NRF1
|
nuclear respiratory factor 1 |
| chr19_-_663171 | 0.15 |
ENST00000606896.1
ENST00000589762.2 |
RNF126
|
ring finger protein 126 |
| chr1_+_153606513 | 0.15 |
ENST00000368694.3
|
CHTOP
|
chromatin target of PRMT1 |
| chr3_+_184098065 | 0.15 |
ENST00000348986.3
|
CHRD
|
chordin |
| chrY_+_2803322 | 0.15 |
ENST00000383052.1
ENST00000155093.3 ENST00000449237.1 ENST00000443793.1 |
ZFY
|
zinc finger protein, Y-linked |
| chr22_-_50746027 | 0.15 |
ENST00000425954.1
ENST00000449103.1 |
PLXNB2
|
plexin B2 |
| chr17_+_80477571 | 0.15 |
ENST00000335255.5
|
FOXK2
|
forkhead box K2 |
| chr11_-_64545941 | 0.15 |
ENST00000377387.1
|
SF1
|
splicing factor 1 |
| chr15_-_72766533 | 0.15 |
ENST00000562573.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr1_+_156698743 | 0.15 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr14_-_74416829 | 0.15 |
ENST00000534936.1
|
FAM161B
|
family with sequence similarity 161, member B |
| chr9_-_131644202 | 0.15 |
ENST00000320665.6
ENST00000436267.2 |
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic |
| chr1_+_110009215 | 0.15 |
ENST00000369872.3
|
SYPL2
|
synaptophysin-like 2 |
| chr9_-_136933615 | 0.15 |
ENST00000371834.2
|
BRD3
|
bromodomain containing 3 |
| chr1_-_43855479 | 0.14 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr13_-_46626820 | 0.14 |
ENST00000428921.1
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr11_+_67033881 | 0.14 |
ENST00000308595.5
ENST00000526285.1 |
ADRBK1
|
adrenergic, beta, receptor kinase 1 |
| chr9_+_34989638 | 0.14 |
ENST00000453597.3
ENST00000335998.3 ENST00000312316.5 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr12_+_50478977 | 0.14 |
ENST00000381513.4
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr3_-_196295437 | 0.14 |
ENST00000429115.1
|
WDR53
|
WD repeat domain 53 |
| chr10_-_17659357 | 0.14 |
ENST00000326961.6
ENST00000361271.3 |
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr17_-_27949911 | 0.14 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr2_+_48541776 | 0.14 |
ENST00000413569.1
ENST00000340553.3 |
FOXN2
|
forkhead box N2 |
| chr14_+_74416989 | 0.14 |
ENST00000334571.2
ENST00000554920.1 |
COQ6
|
coenzyme Q6 monooxygenase |
| chr5_+_10564432 | 0.14 |
ENST00000296657.5
|
ANKRD33B
|
ankyrin repeat domain 33B |
| chr16_+_67226019 | 0.14 |
ENST00000379378.3
|
E2F4
|
E2F transcription factor 4, p107/p130-binding |
| chr5_+_176853702 | 0.13 |
ENST00000507633.1
ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6
|
G protein-coupled receptor kinase 6 |
| chr15_+_91073193 | 0.13 |
ENST00000560098.1
ENST00000268184.6 |
CRTC3
|
CREB regulated transcription coactivator 3 |
| chr11_-_2160180 | 0.13 |
ENST00000381406.4
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr10_-_11653753 | 0.13 |
ENST00000609104.1
|
USP6NL
|
USP6 N-terminal like |
| chr16_-_1823114 | 0.13 |
ENST00000177742.3
ENST00000397375.2 |
MRPS34
|
mitochondrial ribosomal protein S34 |
| chr4_+_53525573 | 0.13 |
ENST00000503051.1
|
USP46-AS1
|
USP46 antisense RNA 1 |
| chr12_-_48213568 | 0.13 |
ENST00000080059.7
ENST00000354334.3 ENST00000430670.1 ENST00000552960.1 ENST00000440293.1 |
HDAC7
|
histone deacetylase 7 |
| chr17_+_48133459 | 0.13 |
ENST00000320031.8
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr10_+_106400859 | 0.13 |
ENST00000369701.3
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr11_+_46354455 | 0.13 |
ENST00000343674.6
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr1_-_19811996 | 0.13 |
ENST00000264203.3
ENST00000401084.2 |
CAPZB
|
capping protein (actin filament) muscle Z-line, beta |
| chr16_+_3019246 | 0.13 |
ENST00000318782.8
ENST00000293978.8 |
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr19_+_4007644 | 0.13 |
ENST00000262971.2
|
PIAS4
|
protein inhibitor of activated STAT, 4 |
| chr3_-_128206759 | 0.13 |
ENST00000430265.2
|
GATA2
|
GATA binding protein 2 |
| chr13_-_80915059 | 0.13 |
ENST00000377104.3
|
SPRY2
|
sprouty homolog 2 (Drosophila) |
| chr3_+_49591881 | 0.13 |
ENST00000296452.4
|
BSN
|
bassoon presynaptic cytomatrix protein |
| chr1_-_19811962 | 0.13 |
ENST00000375142.1
ENST00000264202.6 |
CAPZB
|
capping protein (actin filament) muscle Z-line, beta |
| chr3_-_183979251 | 0.13 |
ENST00000296238.3
|
CAMK2N2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MBD2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.2 | 0.5 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.2 | 0.5 | GO:0061183 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) renal vesicle induction(GO:0072034) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.4 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.5 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.2 | GO:0035283 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.2 | GO:0061386 | noradrenergic neuron differentiation(GO:0003357) closure of optic fissure(GO:0061386) |
| 0.1 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.3 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 0.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.2 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.3 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.4 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.3 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.2 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0048007 | synaptic vesicle recycling via endosome(GO:0036466) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.3 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.2 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0090625 | siRNA loading onto RISC involved in RNA interference(GO:0035087) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.2 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.2 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:2000467 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0090309 | maintenance of DNA methylation(GO:0010216) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.3 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.3 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.2 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.0 | 0.5 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.4 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.0 | 0.1 | GO:1902683 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.0 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.3 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.2 | GO:0002934 | desmosome organization(GO:0002934) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.2 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 0.6 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.4 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.2 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.4 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.2 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.2 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.5 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.6 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 0.7 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |