Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
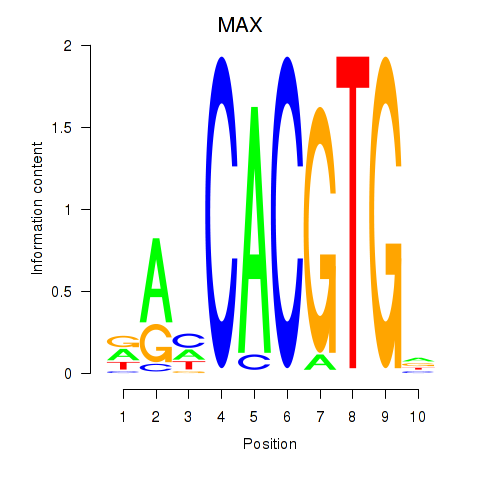
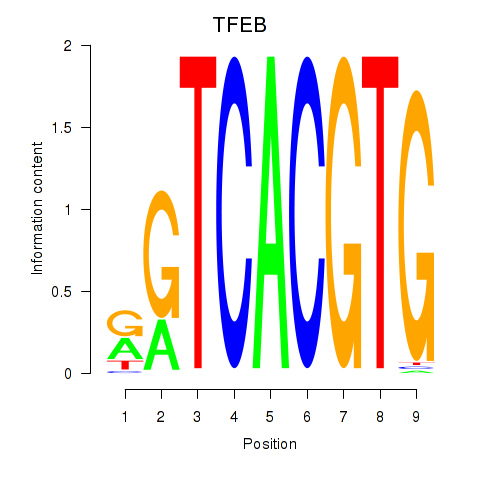
Results for MAX_TFEB
Z-value: 0.48
Transcription factors associated with MAX_TFEB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAX
|
ENSG00000125952.14 | MYC associated factor X |
|
TFEB
|
ENSG00000112561.13 | transcription factor EB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFEB | hg19_v2_chr6_-_41673552_41673678 | 0.78 | 6.7e-02 | Click! |
| MAX | hg19_v2_chr14_-_65569244_65569413 | -0.47 | 3.5e-01 | Click! |
Activity profile of MAX_TFEB motif
Sorted Z-values of MAX_TFEB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_103876528 | 0.86 |
ENST00000522939.1
ENST00000524007.1 |
KB-1507C5.2
|
HCG15011, isoform CRA_a; Protein LOC100996457 |
| chr5_+_133706865 | 0.53 |
ENST00000265339.2
|
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr15_-_65282045 | 0.51 |
ENST00000558765.1
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr6_-_109703663 | 0.49 |
ENST00000368961.5
|
CD164
|
CD164 molecule, sialomucin |
| chrX_-_119693745 | 0.47 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr14_-_54955721 | 0.46 |
ENST00000554908.1
|
GMFB
|
glia maturation factor, beta |
| chr2_+_240323439 | 0.44 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr5_+_133707252 | 0.43 |
ENST00000506787.1
ENST00000507277.1 |
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr5_+_110074685 | 0.41 |
ENST00000355943.3
ENST00000447245.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr10_-_46089939 | 0.41 |
ENST00000453980.3
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr19_+_5681011 | 0.38 |
ENST00000581893.1
ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chr11_-_18343725 | 0.38 |
ENST00000531848.1
|
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr6_-_33385854 | 0.37 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr18_+_9136758 | 0.36 |
ENST00000383440.2
ENST00000262126.4 ENST00000577992.1 |
ANKRD12
|
ankyrin repeat domain 12 |
| chr4_-_47465666 | 0.35 |
ENST00000381571.4
|
COMMD8
|
COMM domain containing 8 |
| chr14_-_54955376 | 0.34 |
ENST00000553333.1
|
GMFB
|
glia maturation factor, beta |
| chr3_+_19988736 | 0.34 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr1_+_52521928 | 0.34 |
ENST00000489308.2
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr8_-_54755789 | 0.33 |
ENST00000359530.2
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr2_-_40006289 | 0.33 |
ENST00000260619.6
ENST00000454352.2 |
THUMPD2
|
THUMP domain containing 2 |
| chr15_-_50978965 | 0.32 |
ENST00000560955.1
ENST00000313478.7 |
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr1_+_42921761 | 0.32 |
ENST00000372562.1
|
PPCS
|
phosphopantothenoylcysteine synthetase |
| chr15_-_65281775 | 0.32 |
ENST00000433215.2
ENST00000558415.1 ENST00000557795.1 |
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr4_-_76439596 | 0.31 |
ENST00000451788.1
ENST00000512706.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr2_-_176867534 | 0.31 |
ENST00000445472.1
|
KIAA1715
|
KIAA1715 |
| chr6_-_109703634 | 0.31 |
ENST00000324953.5
ENST00000310786.4 ENST00000275080.7 ENST00000413644.2 |
CD164
|
CD164 molecule, sialomucin |
| chrX_-_15872914 | 0.31 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr9_+_74526532 | 0.31 |
ENST00000486911.2
|
C9orf85
|
chromosome 9 open reading frame 85 |
| chr3_-_113464906 | 0.31 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr10_+_70883908 | 0.29 |
ENST00000263559.6
ENST00000395098.1 ENST00000546041.1 ENST00000541711.1 |
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe) |
| chr12_+_56110247 | 0.29 |
ENST00000551926.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr9_+_74526384 | 0.29 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr4_-_76439483 | 0.29 |
ENST00000380840.2
ENST00000513257.1 ENST00000507014.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr14_+_77564440 | 0.28 |
ENST00000361786.2
ENST00000555437.1 ENST00000555611.1 ENST00000554658.1 |
KIAA1737
|
CLOCK-interacting pacemaker |
| chr9_-_103115135 | 0.28 |
ENST00000537512.1
|
TEX10
|
testis expressed 10 |
| chr3_-_195163584 | 0.28 |
ENST00000439666.1
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr6_-_84937314 | 0.28 |
ENST00000257766.4
ENST00000403245.3 |
KIAA1009
|
KIAA1009 |
| chr3_+_112280857 | 0.28 |
ENST00000492406.1
ENST00000468642.1 |
SLC35A5
|
solute carrier family 35, member A5 |
| chr14_-_81687197 | 0.28 |
ENST00000553612.1
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr2_-_148778258 | 0.27 |
ENST00000392857.5
ENST00000457954.1 ENST00000392858.1 ENST00000542387.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chrX_+_102585124 | 0.27 |
ENST00000332431.4
ENST00000372666.1 |
TCEAL7
|
transcription elongation factor A (SII)-like 7 |
| chr10_+_23728198 | 0.26 |
ENST00000376495.3
|
OTUD1
|
OTU domain containing 1 |
| chr3_+_184530173 | 0.26 |
ENST00000453056.1
|
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr6_+_7590413 | 0.26 |
ENST00000342415.5
|
SNRNP48
|
small nuclear ribonucleoprotein 48kDa (U11/U12) |
| chr2_-_40006357 | 0.26 |
ENST00000505747.1
|
THUMPD2
|
THUMP domain containing 2 |
| chr4_-_99850243 | 0.25 |
ENST00000280892.6
ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr19_-_49137790 | 0.25 |
ENST00000599385.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr2_+_46926326 | 0.25 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr14_-_35183755 | 0.25 |
ENST00000555765.1
|
CFL2
|
cofilin 2 (muscle) |
| chr6_-_109703600 | 0.25 |
ENST00000512821.1
|
CD164
|
CD164 molecule, sialomucin |
| chr6_-_33385902 | 0.24 |
ENST00000374500.5
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr14_+_64970427 | 0.24 |
ENST00000553583.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr1_+_40627038 | 0.24 |
ENST00000372771.4
|
RLF
|
rearranged L-myc fusion |
| chr1_-_39325431 | 0.24 |
ENST00000373001.3
|
RRAGC
|
Ras-related GTP binding C |
| chr3_-_20227619 | 0.23 |
ENST00000425061.1
ENST00000443724.1 ENST00000421451.1 ENST00000452020.1 ENST00000417364.1 ENST00000306698.2 ENST00000419233.2 ENST00000263753.4 ENST00000383774.1 ENST00000437051.1 ENST00000412868.1 ENST00000429446.3 ENST00000442720.1 |
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chr15_+_82555125 | 0.23 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000566861.1 |
FAM154B
|
family with sequence similarity 154, member B |
| chr17_+_62223320 | 0.23 |
ENST00000580828.1
ENST00000582965.1 |
SNORA76
|
small nucleolar RNA, H/ACA box 76 |
| chr21_-_38445297 | 0.23 |
ENST00000430792.1
ENST00000399103.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr17_-_7137582 | 0.23 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr21_-_38445011 | 0.23 |
ENST00000464265.1
ENST00000399102.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr6_-_80657292 | 0.23 |
ENST00000369816.4
|
ELOVL4
|
ELOVL fatty acid elongase 4 |
| chr8_+_109455830 | 0.23 |
ENST00000524143.1
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr7_+_116593536 | 0.23 |
ENST00000417919.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr4_-_54930790 | 0.22 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr3_-_196987309 | 0.22 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr8_-_54755459 | 0.22 |
ENST00000524234.1
ENST00000521275.1 ENST00000396774.2 |
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr4_+_56719782 | 0.21 |
ENST00000381295.2
ENST00000346134.7 ENST00000349598.6 |
EXOC1
|
exocyst complex component 1 |
| chr13_+_113951607 | 0.21 |
ENST00000397181.3
|
LAMP1
|
lysosomal-associated membrane protein 1 |
| chr17_-_6915646 | 0.21 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
| chr4_+_141264597 | 0.21 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr3_+_127317705 | 0.21 |
ENST00000480910.1
|
MCM2
|
minichromosome maintenance complex component 2 |
| chr12_+_56109926 | 0.21 |
ENST00000547076.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr11_+_67159416 | 0.21 |
ENST00000307980.2
ENST00000544620.1 |
RAD9A
|
RAD9 homolog A (S. pombe) |
| chr4_-_166034004 | 0.21 |
ENST00000505095.1
|
TMEM192
|
transmembrane protein 192 |
| chrX_+_95939638 | 0.20 |
ENST00000373061.3
ENST00000373054.4 ENST00000355827.4 |
DIAPH2
|
diaphanous-related formin 2 |
| chr1_-_183604794 | 0.20 |
ENST00000367534.1
ENST00000359856.6 ENST00000294742.6 |
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa |
| chr9_+_92219919 | 0.20 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr1_+_109419804 | 0.20 |
ENST00000435475.1
|
GPSM2
|
G-protein signaling modulator 2 |
| chr12_+_6833323 | 0.20 |
ENST00000544725.1
|
COPS7A
|
COP9 signalosome subunit 7A |
| chr8_+_9009296 | 0.20 |
ENST00000521718.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr1_+_52521797 | 0.19 |
ENST00000313334.8
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr7_-_108209897 | 0.19 |
ENST00000313516.5
|
THAP5
|
THAP domain containing 5 |
| chr2_+_46926048 | 0.19 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr4_-_84376953 | 0.19 |
ENST00000510985.1
ENST00000295488.3 |
HELQ
|
helicase, POLQ-like |
| chr14_-_35591433 | 0.19 |
ENST00000261475.5
ENST00000555644.1 |
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr18_+_77867177 | 0.18 |
ENST00000560752.1
|
ADNP2
|
ADNP homeobox 2 |
| chr2_-_148778323 | 0.18 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr12_-_76953513 | 0.18 |
ENST00000547540.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr5_+_41904431 | 0.18 |
ENST00000381647.2
|
C5orf51
|
chromosome 5 open reading frame 51 |
| chr17_-_7137857 | 0.18 |
ENST00000005340.5
|
DVL2
|
dishevelled segment polarity protein 2 |
| chr8_+_120220561 | 0.18 |
ENST00000276681.6
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr10_-_93392811 | 0.18 |
ENST00000238994.5
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C |
| chr1_+_222885884 | 0.17 |
ENST00000340934.5
|
BROX
|
BRO1 domain and CAAX motif containing |
| chr1_+_42922173 | 0.17 |
ENST00000455780.1
ENST00000372560.3 ENST00000372561.3 ENST00000372556.3 |
PPCS
|
phosphopantothenoylcysteine synthetase |
| chr10_-_76995675 | 0.17 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr9_+_96928516 | 0.17 |
ENST00000602703.1
|
RP11-2B6.3
|
RP11-2B6.3 |
| chr1_+_100436065 | 0.17 |
ENST00000370153.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr4_+_76439665 | 0.17 |
ENST00000508105.1
ENST00000311638.3 ENST00000380837.3 ENST00000507556.1 ENST00000504190.1 ENST00000507885.1 ENST00000502620.1 ENST00000514480.1 |
THAP6
|
THAP domain containing 6 |
| chr12_+_64845660 | 0.17 |
ENST00000331710.5
|
TBK1
|
TANK-binding kinase 1 |
| chr6_+_123110302 | 0.17 |
ENST00000368440.4
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr1_-_222886526 | 0.17 |
ENST00000541237.1
|
AIDA
|
axin interactor, dorsalization associated |
| chr3_-_142720267 | 0.17 |
ENST00000597953.1
|
RP11-91G21.1
|
RP11-91G21.1 |
| chr3_+_140660634 | 0.16 |
ENST00000446041.2
ENST00000507429.1 ENST00000324194.6 |
SLC25A36
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr8_-_75233563 | 0.16 |
ENST00000342232.4
|
JPH1
|
junctophilin 1 |
| chr2_+_172864490 | 0.16 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr1_-_222885770 | 0.16 |
ENST00000355727.2
ENST00000340020.6 |
AIDA
|
axin interactor, dorsalization associated |
| chr11_+_62538775 | 0.16 |
ENST00000294168.3
ENST00000526261.1 |
TAF6L
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa |
| chr12_-_76953284 | 0.16 |
ENST00000547544.1
ENST00000393249.2 |
OSBPL8
|
oxysterol binding protein-like 8 |
| chr3_-_122512619 | 0.16 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr13_+_42846272 | 0.16 |
ENST00000025301.2
|
AKAP11
|
A kinase (PRKA) anchor protein 11 |
| chr6_+_87865262 | 0.16 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr3_-_57678772 | 0.16 |
ENST00000311128.5
|
DENND6A
|
DENN/MADD domain containing 6A |
| chr18_+_9708162 | 0.16 |
ENST00000578921.1
|
RAB31
|
RAB31, member RAS oncogene family |
| chr3_-_113465065 | 0.15 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr12_-_76477707 | 0.15 |
ENST00000551992.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr1_+_89149905 | 0.15 |
ENST00000316005.7
ENST00000370521.3 ENST00000370505.3 |
PKN2
|
protein kinase N2 |
| chr9_-_6015607 | 0.15 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr11_-_18343669 | 0.15 |
ENST00000396253.3
ENST00000349215.3 ENST00000438420.2 |
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr9_-_123476612 | 0.15 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr10_-_46090334 | 0.15 |
ENST00000395771.3
ENST00000319836.3 |
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr17_-_77967433 | 0.15 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr18_-_29522989 | 0.15 |
ENST00000582539.1
ENST00000283351.4 ENST00000582513.1 |
TRAPPC8
|
trafficking protein particle complex 8 |
| chr11_+_9406169 | 0.15 |
ENST00000379719.3
ENST00000527431.1 |
IPO7
|
importin 7 |
| chr11_-_85780086 | 0.15 |
ENST00000532317.1
ENST00000528256.1 ENST00000526033.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr14_-_35183886 | 0.15 |
ENST00000298159.6
|
CFL2
|
cofilin 2 (muscle) |
| chr15_-_65282274 | 0.15 |
ENST00000204566.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr3_-_101395936 | 0.15 |
ENST00000461821.1
|
ZBTB11
|
zinc finger and BTB domain containing 11 |
| chr6_+_123110465 | 0.15 |
ENST00000539041.1
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr3_-_197025447 | 0.15 |
ENST00000346964.2
ENST00000357674.4 ENST00000314062.3 ENST00000448528.2 ENST00000419553.1 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr7_-_27179814 | 0.15 |
ENST00000522788.1
ENST00000521779.1 |
HOXA3
|
homeobox A3 |
| chrX_-_34675391 | 0.15 |
ENST00000275954.3
|
TMEM47
|
transmembrane protein 47 |
| chr9_-_103115185 | 0.15 |
ENST00000374902.4
|
TEX10
|
testis expressed 10 |
| chr12_+_56109810 | 0.14 |
ENST00000550412.1
ENST00000257899.2 ENST00000548925.1 ENST00000549147.1 |
RP11-644F5.10
BLOC1S1
|
Uncharacterized protein biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr10_-_32217717 | 0.14 |
ENST00000396144.4
ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12
|
Rho GTPase activating protein 12 |
| chr3_+_133292759 | 0.14 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr2_-_238499337 | 0.14 |
ENST00000411462.1
ENST00000409822.1 |
RAB17
|
RAB17, member RAS oncogene family |
| chr6_+_151773583 | 0.14 |
ENST00000545879.1
|
C6orf211
|
chromosome 6 open reading frame 211 |
| chr3_-_197024965 | 0.14 |
ENST00000392382.2
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr1_-_68962744 | 0.14 |
ENST00000525124.1
|
DEPDC1
|
DEP domain containing 1 |
| chr7_+_36429424 | 0.14 |
ENST00000396068.2
|
ANLN
|
anillin, actin binding protein |
| chrX_-_100604184 | 0.14 |
ENST00000372902.3
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr5_+_56205878 | 0.14 |
ENST00000423328.1
|
SETD9
|
SET domain containing 9 |
| chr17_+_49337881 | 0.14 |
ENST00000225298.7
|
UTP18
|
UTP18 small subunit (SSU) processome component homolog (yeast) |
| chr17_-_49337392 | 0.14 |
ENST00000376381.2
ENST00000586178.1 |
MBTD1
|
mbt domain containing 1 |
| chr3_-_45883558 | 0.14 |
ENST00000445698.1
ENST00000296135.6 |
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chrX_+_150565038 | 0.14 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr1_+_15853308 | 0.14 |
ENST00000375838.1
ENST00000375847.3 ENST00000375849.1 |
DNAJC16
|
DnaJ (Hsp40) homolog, subfamily C, member 16 |
| chr5_+_110427983 | 0.13 |
ENST00000513710.2
ENST00000505303.1 |
WDR36
|
WD repeat domain 36 |
| chr2_+_232575168 | 0.13 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr16_-_81129845 | 0.13 |
ENST00000569885.1
ENST00000566566.1 |
GCSH
|
glycine cleavage system protein H (aminomethyl carrier) |
| chr14_-_54420133 | 0.13 |
ENST00000559501.1
ENST00000558984.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr3_+_140660743 | 0.13 |
ENST00000453248.2
|
SLC25A36
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr14_-_35591156 | 0.13 |
ENST00000554361.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr3_+_19988566 | 0.13 |
ENST00000273047.4
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chrX_+_95939711 | 0.13 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr18_+_20715416 | 0.13 |
ENST00000580153.1
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr21_+_38445539 | 0.13 |
ENST00000418766.1
ENST00000450533.1 ENST00000438055.1 ENST00000355666.1 ENST00000540756.1 ENST00000399010.1 |
TTC3
|
tetratricopeptide repeat domain 3 |
| chr17_-_19880992 | 0.13 |
ENST00000395536.3
ENST00000576896.1 ENST00000225737.6 |
AKAP10
|
A kinase (PRKA) anchor protein 10 |
| chr11_-_113746212 | 0.13 |
ENST00000537642.1
ENST00000537706.1 ENST00000544750.1 ENST00000260188.5 ENST00000540925.1 |
USP28
|
ubiquitin specific peptidase 28 |
| chr13_-_79233314 | 0.13 |
ENST00000282003.6
|
RNF219
|
ring finger protein 219 |
| chr6_-_33385823 | 0.13 |
ENST00000494751.1
ENST00000374496.3 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr8_-_95961578 | 0.13 |
ENST00000448464.2
ENST00000342697.4 |
TP53INP1
|
tumor protein p53 inducible nuclear protein 1 |
| chr6_+_139349817 | 0.13 |
ENST00000367660.3
|
ABRACL
|
ABRA C-terminal like |
| chr7_+_92861653 | 0.13 |
ENST00000251739.5
ENST00000305866.5 ENST00000544910.1 ENST00000541136.1 ENST00000458530.1 ENST00000535481.1 ENST00000317751.6 |
CCDC132
|
coiled-coil domain containing 132 |
| chr8_+_104426942 | 0.13 |
ENST00000297579.5
|
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr10_+_112327425 | 0.13 |
ENST00000361804.4
|
SMC3
|
structural maintenance of chromosomes 3 |
| chr9_-_123476719 | 0.13 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr2_-_55459485 | 0.13 |
ENST00000451916.1
|
CLHC1
|
clathrin heavy chain linker domain containing 1 |
| chr20_+_17550691 | 0.13 |
ENST00000474024.1
|
DSTN
|
destrin (actin depolymerizing factor) |
| chr12_+_28343353 | 0.12 |
ENST00000539107.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr1_-_71546690 | 0.12 |
ENST00000254821.6
|
ZRANB2
|
zinc finger, RAN-binding domain containing 2 |
| chr12_-_42632016 | 0.12 |
ENST00000442791.3
ENST00000327791.4 ENST00000534854.2 ENST00000380788.3 ENST00000380790.4 |
YAF2
|
YY1 associated factor 2 |
| chr19_-_11545920 | 0.12 |
ENST00000356392.4
ENST00000591179.1 |
CCDC151
|
coiled-coil domain containing 151 |
| chr12_-_108154705 | 0.12 |
ENST00000547188.1
|
PRDM4
|
PR domain containing 4 |
| chr7_+_108210012 | 0.12 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr22_-_36903101 | 0.12 |
ENST00000397224.4
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr17_+_42977122 | 0.12 |
ENST00000412523.2
ENST00000331733.4 ENST00000417826.2 |
FAM187A
CCDC103
|
family with sequence similarity 187, member A coiled-coil domain containing 103 |
| chr5_-_176730733 | 0.12 |
ENST00000504395.1
|
RAB24
|
RAB24, member RAS oncogene family |
| chrX_+_72783026 | 0.12 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr11_+_18344106 | 0.12 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr11_+_114310237 | 0.12 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chr7_+_36429409 | 0.12 |
ENST00000265748.2
|
ANLN
|
anillin, actin binding protein |
| chr1_+_17248418 | 0.12 |
ENST00000375541.5
|
CROCC
|
ciliary rootlet coiled-coil, rootletin |
| chr2_-_198364581 | 0.12 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chrX_-_102531717 | 0.12 |
ENST00000372680.1
|
TCEAL5
|
transcription elongation factor A (SII)-like 5 |
| chr11_+_120894781 | 0.12 |
ENST00000529397.1
ENST00000528512.1 ENST00000422003.2 |
TBCEL
|
tubulin folding cofactor E-like |
| chr17_-_57184260 | 0.12 |
ENST00000376149.3
ENST00000393066.3 |
TRIM37
|
tripartite motif containing 37 |
| chr19_+_8386371 | 0.12 |
ENST00000600659.2
|
RPS28
|
ribosomal protein S28 |
| chr7_+_12726474 | 0.12 |
ENST00000396662.1
ENST00000356797.3 ENST00000396664.2 |
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr22_-_42343117 | 0.12 |
ENST00000407253.3
ENST00000215980.5 |
CENPM
|
centromere protein M |
| chr16_+_24550857 | 0.12 |
ENST00000568015.1
|
RBBP6
|
retinoblastoma binding protein 6 |
| chr7_+_74379083 | 0.12 |
ENST00000361825.7
|
GATSL1
|
GATS protein-like 1 |
| chr11_-_47470682 | 0.11 |
ENST00000529341.1
ENST00000352508.3 |
RAPSN
|
receptor-associated protein of the synapse |
| chr12_+_64798095 | 0.11 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr9_+_130860583 | 0.11 |
ENST00000373064.5
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr3_+_50654821 | 0.11 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr16_+_57220193 | 0.11 |
ENST00000564435.1
ENST00000562959.1 ENST00000394420.4 ENST00000568505.2 ENST00000537866.1 |
RSPRY1
|
ring finger and SPRY domain containing 1 |
| chr12_-_46663734 | 0.11 |
ENST00000550173.1
|
SLC38A1
|
solute carrier family 38, member 1 |
| chr17_-_18266818 | 0.11 |
ENST00000583780.1
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr11_+_107879459 | 0.11 |
ENST00000393094.2
|
CUL5
|
cullin 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAX_TFEB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.2 | 0.2 | GO:0010737 | protein kinase A signaling(GO:0010737) regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.3 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 | 0.4 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.1 | 0.4 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.8 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.3 | GO:0031106 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.1 | 0.3 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.1 | 0.4 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 0.5 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.6 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.3 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.2 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.1 | 0.3 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.4 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0045041 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.2 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.5 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.0 | 0.7 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.1 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.0 | 0.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.0 | 0.1 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.0 | 0.3 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.2 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0072096 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.7 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.8 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.1 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.0 | 0.1 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.2 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:0042414 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) epinephrine metabolic process(GO:0042414) cellular response to lead ion(GO:0071284) |
| 0.0 | 0.1 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.6 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.2 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.0 | GO:1903371 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.2 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 1.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.9 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.0 | GO:0045575 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.0 | 0.0 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.0 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.6 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.2 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.3 | GO:0036010 | protein localization to endosome(GO:0036010) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.8 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 1.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.6 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.3 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 1.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) omegasome membrane(GO:1903349) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.0 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.0 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.2 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.2 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 1.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0000248 | C-5 sterol desaturase activity(GO:0000248) sterol desaturase activity(GO:0070704) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.0 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |