Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
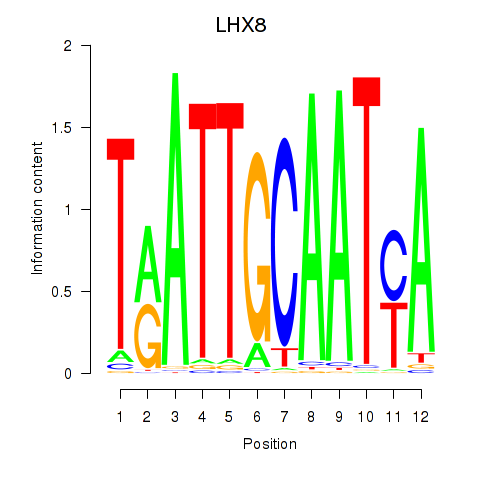
Results for LHX8
Z-value: 0.59
Transcription factors associated with LHX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX8
|
ENSG00000162624.10 | LIM homeobox 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX8 | hg19_v2_chr1_+_75600567_75600567 | 0.67 | 1.5e-01 | Click! |
Activity profile of LHX8 motif
Sorted Z-values of LHX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_21688366 | 0.53 |
ENST00000358491.4
ENST00000597078.1 |
ZNF429
|
zinc finger protein 429 |
| chr17_+_58018269 | 0.53 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr1_+_109419804 | 0.46 |
ENST00000435475.1
|
GPSM2
|
G-protein signaling modulator 2 |
| chr3_+_119187785 | 0.41 |
ENST00000295588.4
ENST00000476573.1 |
POGLUT1
|
protein O-glucosyltransferase 1 |
| chr18_-_14132422 | 0.39 |
ENST00000589498.1
ENST00000590202.1 |
ZNF519
|
zinc finger protein 519 |
| chr8_+_9009296 | 0.29 |
ENST00000521718.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr8_+_101349823 | 0.29 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr1_+_95616933 | 0.28 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr2_+_171640291 | 0.27 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr10_-_70231639 | 0.27 |
ENST00000551118.2
ENST00000358410.3 ENST00000399180.2 ENST00000399179.2 |
DNA2
|
DNA replication helicase/nuclease 2 |
| chr12_-_15038779 | 0.25 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr2_-_136678123 | 0.25 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr6_-_56507586 | 0.25 |
ENST00000439203.1
ENST00000518935.1 ENST00000446842.2 ENST00000370765.6 ENST00000244364.6 |
DST
|
dystonin |
| chr4_-_36245561 | 0.24 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr5_-_24645078 | 0.24 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr1_-_168464875 | 0.24 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr19_-_58204128 | 0.23 |
ENST00000597520.1
|
AC004017.1
|
Uncharacterized protein |
| chr1_+_144989309 | 0.23 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr9_+_26956371 | 0.22 |
ENST00000380062.5
ENST00000518614.1 |
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr2_-_178128250 | 0.21 |
ENST00000448782.1
ENST00000446151.2 |
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr22_-_29107919 | 0.21 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr7_+_12727250 | 0.21 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr17_+_48823975 | 0.21 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr2_-_152118352 | 0.20 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr8_-_82608409 | 0.20 |
ENST00000518568.1
|
SLC10A5
|
solute carrier family 10, member 5 |
| chr10_-_28571015 | 0.20 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr1_-_110933663 | 0.19 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr2_+_102758271 | 0.19 |
ENST00000428279.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr3_+_127317945 | 0.19 |
ENST00000472731.1
|
MCM2
|
minichromosome maintenance complex component 2 |
| chrX_-_117119243 | 0.18 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chrX_+_56258844 | 0.18 |
ENST00000374928.3
|
KLF8
|
Kruppel-like factor 8 |
| chr1_-_236445251 | 0.17 |
ENST00000354619.5
ENST00000327333.8 |
ERO1LB
|
ERO1-like beta (S. cerevisiae) |
| chr1_-_149459549 | 0.17 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr10_+_94352956 | 0.17 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr1_-_110933611 | 0.17 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr4_+_174089904 | 0.16 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr18_-_11908272 | 0.16 |
ENST00000592977.1
ENST00000590501.1 ENST00000586844.1 |
MPPE1
|
metallophosphoesterase 1 |
| chr6_+_154360616 | 0.16 |
ENST00000229768.5
ENST00000419506.2 ENST00000524163.1 ENST00000414028.2 ENST00000435918.2 ENST00000337049.4 |
OPRM1
|
opioid receptor, mu 1 |
| chr4_+_166300084 | 0.16 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr7_+_30589829 | 0.15 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr4_-_69111401 | 0.15 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr19_+_21264980 | 0.15 |
ENST00000596053.1
ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714
|
zinc finger protein 714 |
| chr11_-_118122996 | 0.15 |
ENST00000525386.1
ENST00000527472.1 ENST00000278949.4 |
MPZL3
|
myelin protein zero-like 3 |
| chr2_-_178128149 | 0.14 |
ENST00000423513.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr11_+_125757556 | 0.14 |
ENST00000526028.1
|
HYLS1
|
hydrolethalus syndrome 1 |
| chr2_+_200775971 | 0.14 |
ENST00000319974.5
|
C2orf69
|
chromosome 2 open reading frame 69 |
| chr9_+_26956474 | 0.13 |
ENST00000429045.2
|
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr2_-_152830441 | 0.13 |
ENST00000534999.1
ENST00000397327.2 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr2_+_216946589 | 0.13 |
ENST00000433112.1
ENST00000454545.1 ENST00000437356.2 ENST00000295658.4 ENST00000455479.1 ENST00000406027.2 |
TMEM169
|
transmembrane protein 169 |
| chr2_-_178128528 | 0.13 |
ENST00000397063.4
ENST00000421929.1 |
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr2_+_217363793 | 0.13 |
ENST00000456586.1
ENST00000598925.1 ENST00000427280.2 |
RPL37A
|
ribosomal protein L37a |
| chr16_-_53737722 | 0.13 |
ENST00000569716.1
ENST00000562588.1 ENST00000562230.1 ENST00000379925.3 ENST00000563746.1 ENST00000568653.3 |
RPGRIP1L
|
RPGRIP1-like |
| chr10_-_47151341 | 0.12 |
ENST00000422732.2
|
LINC00842
|
long intergenic non-protein coding RNA 842 |
| chr4_-_153332886 | 0.12 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr5_-_130868688 | 0.12 |
ENST00000504575.1
ENST00000513227.1 |
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr12_-_88974236 | 0.12 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr2_-_88285309 | 0.12 |
ENST00000420840.2
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr14_+_57046500 | 0.12 |
ENST00000261556.6
|
TMEM260
|
transmembrane protein 260 |
| chr9_-_13165457 | 0.12 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr19_+_44716678 | 0.12 |
ENST00000586228.1
ENST00000588219.1 ENST00000313040.7 ENST00000589707.1 ENST00000588394.1 ENST00000589005.1 |
ZNF227
|
zinc finger protein 227 |
| chr5_+_157158205 | 0.12 |
ENST00000231198.7
|
THG1L
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr19_+_44716768 | 0.12 |
ENST00000586048.1
|
ZNF227
|
zinc finger protein 227 |
| chr19_-_4535233 | 0.12 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr5_-_39270725 | 0.12 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr4_+_174089951 | 0.12 |
ENST00000512285.1
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr2_+_190541153 | 0.11 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chrX_+_102883620 | 0.11 |
ENST00000372626.3
|
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr6_+_26087509 | 0.11 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr6_+_26087646 | 0.11 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr3_+_154798162 | 0.11 |
ENST00000360490.2
|
MME
|
membrane metallo-endopeptidase |
| chr12_+_97306295 | 0.11 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr14_+_50291993 | 0.11 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr2_+_201994208 | 0.11 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr3_+_42977846 | 0.10 |
ENST00000383748.4
|
KRBOX1
|
KRAB box domain containing 1 |
| chr15_-_40074996 | 0.10 |
ENST00000350221.3
|
FSIP1
|
fibrous sheath interacting protein 1 |
| chr2_+_86947296 | 0.10 |
ENST00000283632.4
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr2_+_102758210 | 0.10 |
ENST00000450319.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr12_+_48516463 | 0.10 |
ENST00000546465.1
|
PFKM
|
phosphofructokinase, muscle |
| chr9_+_70856899 | 0.09 |
ENST00000377342.5
ENST00000478048.1 |
CBWD3
|
COBW domain containing 3 |
| chr1_+_92632542 | 0.09 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr2_+_87135076 | 0.09 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr2_+_87144738 | 0.09 |
ENST00000559485.1
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr11_-_3400442 | 0.09 |
ENST00000429541.2
ENST00000532539.1 |
ZNF195
|
zinc finger protein 195 |
| chr9_-_5833027 | 0.09 |
ENST00000339450.5
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr1_+_160370344 | 0.09 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr11_+_59480899 | 0.09 |
ENST00000300150.7
|
STX3
|
syntaxin 3 |
| chr1_-_146040968 | 0.09 |
ENST00000401010.3
|
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr16_-_53737795 | 0.09 |
ENST00000262135.4
ENST00000564374.1 ENST00000566096.1 |
RPGRIP1L
|
RPGRIP1-like |
| chr18_-_11908329 | 0.08 |
ENST00000344987.7
ENST00000588103.1 ENST00000588191.1 ENST00000317235.7 ENST00000309976.9 ENST00000588186.1 ENST00000589267.1 |
MPPE1
|
metallophosphoesterase 1 |
| chr16_-_15149828 | 0.08 |
ENST00000566419.1
ENST00000568320.1 |
NTAN1
|
N-terminal asparagine amidase |
| chr2_-_178128199 | 0.08 |
ENST00000449627.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr1_-_212004090 | 0.08 |
ENST00000366997.4
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr16_-_15149917 | 0.08 |
ENST00000287706.3
|
NTAN1
|
N-terminal asparagine amidase |
| chr2_-_37068530 | 0.08 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr8_-_18711866 | 0.08 |
ENST00000519851.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr7_-_22234381 | 0.08 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr4_-_100242549 | 0.08 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr7_-_20256965 | 0.07 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr12_+_9822331 | 0.07 |
ENST00000545918.1
ENST00000543300.1 ENST00000261339.6 ENST00000466035.2 |
CLEC2D
|
C-type lectin domain family 2, member D |
| chr4_-_186125077 | 0.07 |
ENST00000458385.2
ENST00000514798.1 ENST00000296775.6 |
KIAA1430
|
KIAA1430 |
| chr14_+_39583427 | 0.07 |
ENST00000308317.6
ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2
|
gem (nuclear organelle) associated protein 2 |
| chr7_+_115862858 | 0.07 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr12_+_26205496 | 0.07 |
ENST00000537946.1
ENST00000541218.1 ENST00000282884.9 ENST00000545413.1 |
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr1_+_113009163 | 0.07 |
ENST00000256640.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr2_-_225266802 | 0.07 |
ENST00000243806.2
|
FAM124B
|
family with sequence similarity 124B |
| chr2_+_114195268 | 0.07 |
ENST00000259199.4
ENST00000416503.2 ENST00000433343.2 |
CBWD2
|
COBW domain containing 2 |
| chr7_-_6865826 | 0.07 |
ENST00000538180.1
|
CCZ1B
|
CCZ1 vacuolar protein trafficking and biogenesis associated homolog B (S. cerevisiae) |
| chr1_-_48937838 | 0.07 |
ENST00000371847.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr7_+_55980331 | 0.07 |
ENST00000429591.2
|
ZNF713
|
zinc finger protein 713 |
| chr19_+_21203426 | 0.07 |
ENST00000261560.5
ENST00000599548.1 ENST00000594110.1 |
ZNF430
|
zinc finger protein 430 |
| chr15_+_78832747 | 0.07 |
ENST00000560217.1
ENST00000044462.7 ENST00000559082.1 ENST00000559948.1 ENST00000413382.2 ENST00000559146.1 ENST00000558281.1 |
PSMA4
|
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr3_+_195447738 | 0.07 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr5_+_41904431 | 0.06 |
ENST00000381647.2
|
C5orf51
|
chromosome 5 open reading frame 51 |
| chr13_+_53216565 | 0.06 |
ENST00000357495.2
|
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2 |
| chr4_+_106631966 | 0.06 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr7_+_151791074 | 0.06 |
ENST00000447796.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr7_+_5938386 | 0.06 |
ENST00000537980.1
|
CCZ1
|
CCZ1 vacuolar protein trafficking and biogenesis associated homolog (S. cerevisiae) |
| chr8_-_62602327 | 0.06 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr1_+_53662101 | 0.06 |
ENST00000371486.3
|
CPT2
|
carnitine palmitoyltransferase 2 |
| chr17_-_42144949 | 0.06 |
ENST00000591247.1
|
LSM12
|
LSM12 homolog (S. cerevisiae) |
| chr9_-_21974820 | 0.06 |
ENST00000579122.1
ENST00000498124.1 |
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr6_+_116691001 | 0.06 |
ENST00000537543.1
|
DSE
|
dermatan sulfate epimerase |
| chr7_-_140482926 | 0.06 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chrX_+_13707235 | 0.06 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr8_+_75262612 | 0.06 |
ENST00000220822.7
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr9_+_70856397 | 0.06 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr15_+_65843130 | 0.06 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr3_-_187455680 | 0.06 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr8_+_22424551 | 0.06 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr14_-_92247032 | 0.06 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr5_+_135170331 | 0.06 |
ENST00000425402.1
ENST00000274513.5 ENST00000420621.1 ENST00000433282.2 ENST00000412661.2 |
SLC25A48
|
solute carrier family 25, member 48 |
| chr19_+_12175504 | 0.06 |
ENST00000439326.3
|
ZNF844
|
zinc finger protein 844 |
| chr4_+_74718906 | 0.05 |
ENST00000226524.3
|
PF4V1
|
platelet factor 4 variant 1 |
| chr5_+_67088974 | 0.05 |
ENST00000513234.1
ENST00000504068.1 ENST00000515227.1 |
RP11-434D9.1
|
RP11-434D9.1 |
| chr11_+_6226782 | 0.05 |
ENST00000316375.2
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr20_+_30458431 | 0.05 |
ENST00000375938.4
ENST00000535842.1 ENST00000310998.4 ENST00000375921.2 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr19_+_42300548 | 0.05 |
ENST00000344550.4
|
CEACAM3
|
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr4_-_110723335 | 0.05 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr1_-_48937821 | 0.05 |
ENST00000396199.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr13_+_38923959 | 0.05 |
ENST00000379649.1
ENST00000239878.4 ENST00000437952.1 ENST00000379641.1 |
UFM1
|
ubiquitin-fold modifier 1 |
| chr1_+_146373546 | 0.05 |
ENST00000446760.2
|
NBPF12
|
neuroblastoma breakpoint family, member 12 |
| chr16_-_11922665 | 0.05 |
ENST00000573319.1
ENST00000577041.1 ENST00000574028.1 ENST00000571259.1 ENST00000573037.1 ENST00000571158.1 |
BCAR4
|
breast cancer anti-estrogen resistance 4 (non-protein coding) |
| chr4_+_57302297 | 0.05 |
ENST00000399688.3
ENST00000512576.1 |
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr1_+_145883868 | 0.05 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr11_-_68780824 | 0.05 |
ENST00000441623.1
ENST00000309099.6 |
MRGPRF
|
MAS-related GPR, member F |
| chr7_+_151791095 | 0.05 |
ENST00000422997.2
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr12_+_45686457 | 0.04 |
ENST00000441606.2
|
ANO6
|
anoctamin 6 |
| chr9_-_179018 | 0.04 |
ENST00000431099.2
ENST00000382447.4 ENST00000382389.1 ENST00000377447.3 ENST00000314367.10 ENST00000356521.4 ENST00000382393.1 ENST00000377400.4 |
CBWD1
|
COBW domain containing 1 |
| chr2_+_234621551 | 0.04 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr15_-_34610962 | 0.04 |
ENST00000290209.5
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr6_-_4347271 | 0.04 |
ENST00000437430.2
|
RP3-527G5.1
|
RP3-527G5.1 |
| chr11_+_29181503 | 0.04 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr6_+_43543942 | 0.04 |
ENST00000372226.1
ENST00000443535.1 |
POLH
|
polymerase (DNA directed), eta |
| chr2_-_152830479 | 0.04 |
ENST00000360283.6
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr2_+_158114051 | 0.04 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr7_-_28220354 | 0.04 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr10_+_90672113 | 0.04 |
ENST00000371922.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr6_+_153071925 | 0.04 |
ENST00000367244.3
ENST00000367243.3 |
VIP
|
vasoactive intestinal peptide |
| chr4_-_95264008 | 0.04 |
ENST00000295256.5
|
HPGDS
|
hematopoietic prostaglandin D synthase |
| chr1_+_202789394 | 0.03 |
ENST00000330493.5
|
RP11-480I12.4
|
Putative inactive alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase-like protein LOC641515 |
| chr1_+_145293371 | 0.03 |
ENST00000342960.5
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr12_-_11548496 | 0.03 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr2_+_128377550 | 0.03 |
ENST00000437387.1
ENST00000409090.1 |
MYO7B
|
myosin VIIB |
| chrX_-_21776281 | 0.03 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr7_-_7575477 | 0.03 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr1_-_160492994 | 0.03 |
ENST00000368055.1
ENST00000368057.3 ENST00000368059.3 |
SLAMF6
|
SLAM family member 6 |
| chr1_-_95391315 | 0.03 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr11_-_104817919 | 0.03 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chrX_+_70798261 | 0.03 |
ENST00000373696.3
|
ACRC
|
acidic repeat containing |
| chr4_+_26344754 | 0.03 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr2_-_240969906 | 0.03 |
ENST00000402971.2
|
OR6B2
|
olfactory receptor, family 6, subfamily B, member 2 |
| chr4_-_110723194 | 0.03 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr14_+_23067166 | 0.03 |
ENST00000216327.6
ENST00000542041.1 |
ABHD4
|
abhydrolase domain containing 4 |
| chr13_+_96085847 | 0.03 |
ENST00000376873.3
|
CLDN10
|
claudin 10 |
| chrX_-_20236970 | 0.03 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr7_+_151791037 | 0.03 |
ENST00000419245.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr16_-_88770019 | 0.03 |
ENST00000541206.2
|
RNF166
|
ring finger protein 166 |
| chr15_-_65426174 | 0.03 |
ENST00000204549.4
|
PDCD7
|
programmed cell death 7 |
| chr6_+_10633993 | 0.02 |
ENST00000417671.1
|
GCNT6
|
glucosaminyl (N-acetyl) transferase 6 |
| chr1_+_70876926 | 0.02 |
ENST00000370938.3
ENST00000346806.2 |
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr1_+_47603109 | 0.02 |
ENST00000371890.3
ENST00000294337.3 ENST00000371891.3 |
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chr4_-_83812248 | 0.02 |
ENST00000514326.1
ENST00000505434.1 ENST00000503058.1 ENST00000348405.4 ENST00000505984.1 ENST00000513858.1 ENST00000508479.1 ENST00000443462.2 ENST00000508502.1 ENST00000509142.1 ENST00000432794.1 ENST00000448323.1 ENST00000326950.5 ENST00000311785.7 |
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr22_-_29457832 | 0.02 |
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr18_-_74839891 | 0.02 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr14_+_62164340 | 0.02 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr9_+_130160440 | 0.02 |
ENST00000439597.1
ENST00000423934.1 |
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr5_+_140165876 | 0.02 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr4_-_83812402 | 0.02 |
ENST00000395310.2
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr6_+_160221293 | 0.02 |
ENST00000610273.1
ENST00000392167.3 |
PNLDC1
|
poly(A)-specific ribonuclease (PARN)-like domain containing 1 |
| chr1_+_45241109 | 0.02 |
ENST00000396651.3
|
RPS8
|
ribosomal protein S8 |
| chr15_-_75918787 | 0.02 |
ENST00000564086.1
|
SNUPN
|
snurportin 1 |
| chr6_+_76599809 | 0.02 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr15_-_75918500 | 0.02 |
ENST00000569817.1
|
SNUPN
|
snurportin 1 |
| chr1_+_224301787 | 0.02 |
ENST00000366862.5
ENST00000424254.2 |
FBXO28
|
F-box protein 28 |
| chrX_+_73164167 | 0.02 |
ENST00000414209.1
ENST00000602895.1 ENST00000453317.1 ENST00000602546.1 ENST00000602985.1 ENST00000415215.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr16_-_20338748 | 0.02 |
ENST00000575582.1
ENST00000341642.5 ENST00000381362.4 ENST00000572347.1 ENST00000572478.1 ENST00000302555.5 |
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr2_-_136288113 | 0.02 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr4_-_48082192 | 0.02 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr13_-_37633567 | 0.01 |
ENST00000464744.1
|
SUPT20H
|
suppressor of Ty 20 homolog (S. cerevisiae) |
| chr5_+_175511859 | 0.01 |
ENST00000503724.2
ENST00000253490.4 |
FAM153B
|
family with sequence similarity 153, member B |
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.1 | 0.5 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 0.6 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.1 | 0.2 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.1 | 0.2 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.3 | GO:1902990 | DNA replication, removal of RNA primer(GO:0043137) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.4 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.3 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.1 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.2 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:1903464 | negative regulation of mitotic cell cycle DNA replication(GO:1903464) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.2 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.3 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |