Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for LHX2
Z-value: 0.49
Transcription factors associated with LHX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX2
|
ENSG00000106689.6 | LIM homeobox 2 |
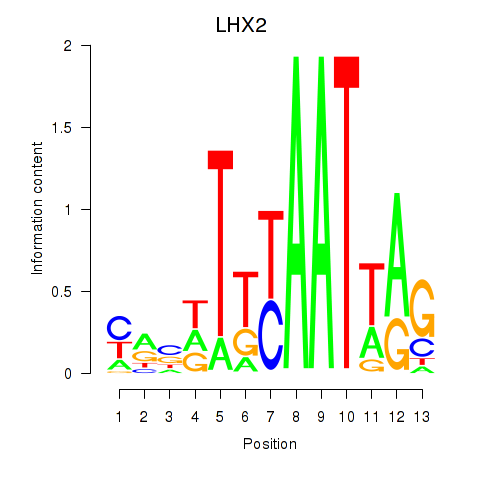
Activity profile of LHX2 motif
Sorted Z-values of LHX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_26199471 | 0.35 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr17_-_7760457 | 0.26 |
ENST00000576384.1
|
LSMD1
|
LSM domain containing 1 |
| chr6_-_26199499 | 0.25 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr2_-_166702601 | 0.25 |
ENST00000428888.1
|
AC009495.4
|
AC009495.4 |
| chr1_-_168464875 | 0.24 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr1_+_219347203 | 0.21 |
ENST00000366927.3
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr2_-_160568529 | 0.20 |
ENST00000418770.1
|
AC009961.3
|
AC009961.3 |
| chr7_-_99716914 | 0.19 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr21_-_43735628 | 0.18 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr2_+_145780767 | 0.18 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr11_-_62323702 | 0.17 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr12_+_122688090 | 0.17 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr1_+_84630574 | 0.15 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr2_-_55237484 | 0.14 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr7_-_54826869 | 0.14 |
ENST00000450622.1
|
SEC61G
|
Sec61 gamma subunit |
| chr12_+_41136144 | 0.13 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr16_+_53412368 | 0.13 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr2_-_55276320 | 0.12 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr1_+_192127578 | 0.12 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr7_-_54826920 | 0.11 |
ENST00000395535.3
ENST00000352861.4 |
SEC61G
|
Sec61 gamma subunit |
| chr8_+_22424551 | 0.11 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr15_-_42565023 | 0.10 |
ENST00000566474.1
|
TMEM87A
|
transmembrane protein 87A |
| chr18_+_32556892 | 0.10 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr7_+_37960163 | 0.10 |
ENST00000199448.4
ENST00000559325.1 ENST00000423717.1 |
EPDR1
|
ependymin related 1 |
| chr16_-_66864806 | 0.10 |
ENST00000566336.1
ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr12_-_100656134 | 0.09 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr4_-_185694872 | 0.09 |
ENST00000505492.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr4_-_105416039 | 0.09 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr18_+_3466248 | 0.09 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr2_+_109204909 | 0.09 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr13_+_19756173 | 0.08 |
ENST00000382988.2
|
RP11-408E5.4
|
RP11-408E5.4 |
| chr13_-_51101468 | 0.08 |
ENST00000428276.1
|
RP11-175B12.2
|
RP11-175B12.2 |
| chr19_+_42212526 | 0.08 |
ENST00000598976.1
ENST00000435837.2 ENST00000221992.6 ENST00000405816.1 |
CEA
CEACAM5
|
Uncharacterized protein carcinoembryonic antigen-related cell adhesion molecule 5 |
| chrX_-_138724677 | 0.08 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr18_+_29027696 | 0.08 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr5_+_68860949 | 0.08 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr11_+_27076764 | 0.07 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr21_-_43735446 | 0.07 |
ENST00000398431.2
|
TFF3
|
trefoil factor 3 (intestinal) |
| chr7_-_99717463 | 0.07 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr2_-_175712270 | 0.07 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr1_+_62439037 | 0.07 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr19_-_44384291 | 0.07 |
ENST00000324394.6
|
ZNF404
|
zinc finger protein 404 |
| chr3_+_38017264 | 0.07 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr9_+_108320392 | 0.06 |
ENST00000602661.1
ENST00000223528.2 ENST00000448551.2 ENST00000540160.1 |
FKTN
|
fukutin |
| chr18_-_44702668 | 0.06 |
ENST00000256433.3
|
IER3IP1
|
immediate early response 3 interacting protein 1 |
| chr4_-_89951028 | 0.06 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr10_-_90712520 | 0.06 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr8_+_76452097 | 0.06 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr2_+_29001711 | 0.06 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr7_-_99716952 | 0.06 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr14_-_38036271 | 0.06 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr9_-_77643307 | 0.06 |
ENST00000376834.3
ENST00000376830.3 |
C9orf41
|
chromosome 9 open reading frame 41 |
| chr7_-_22234381 | 0.06 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr1_+_160709055 | 0.05 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr18_+_66465302 | 0.05 |
ENST00000360242.5
ENST00000358653.5 |
CCDC102B
|
coiled-coil domain containing 102B |
| chr12_-_118796910 | 0.05 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr1_-_147142358 | 0.05 |
ENST00000392988.2
|
ACP6
|
acid phosphatase 6, lysophosphatidic |
| chr11_-_4719072 | 0.05 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr4_-_70626314 | 0.05 |
ENST00000510821.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr12_+_94071129 | 0.05 |
ENST00000552983.1
ENST00000332896.3 ENST00000552033.1 ENST00000548483.1 |
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr10_+_96443204 | 0.05 |
ENST00000339022.5
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr7_+_80253387 | 0.05 |
ENST00000438020.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr14_+_67708344 | 0.05 |
ENST00000557237.1
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr9_+_116207007 | 0.05 |
ENST00000374140.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr14_+_56078695 | 0.05 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr8_+_92114060 | 0.05 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr19_+_42212501 | 0.05 |
ENST00000398599.4
|
CEACAM5
|
carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr12_+_64173583 | 0.04 |
ENST00000261234.6
|
TMEM5
|
transmembrane protein 5 |
| chr11_+_24518723 | 0.04 |
ENST00000336930.6
ENST00000529015.1 ENST00000533227.1 |
LUZP2
|
leucine zipper protein 2 |
| chr12_-_91573249 | 0.04 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr13_+_78315528 | 0.04 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr11_+_31391381 | 0.04 |
ENST00000465995.1
ENST00000536040.1 |
DNAJC24
|
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr9_+_21440440 | 0.04 |
ENST00000276927.1
|
IFNA1
|
interferon, alpha 1 |
| chr11_+_63606373 | 0.04 |
ENST00000402010.2
ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr13_+_24844857 | 0.04 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr11_-_104972158 | 0.04 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr6_+_125524785 | 0.04 |
ENST00000392482.2
|
TPD52L1
|
tumor protein D52-like 1 |
| chrX_-_20074895 | 0.04 |
ENST00000543767.1
|
MAP7D2
|
MAP7 domain containing 2 |
| chr14_+_100532771 | 0.04 |
ENST00000557153.1
|
EVL
|
Enah/Vasp-like |
| chr6_+_53883708 | 0.04 |
ENST00000514921.1
ENST00000274897.5 ENST00000370877.2 |
MLIP
|
muscular LMNA-interacting protein |
| chr14_-_23395623 | 0.04 |
ENST00000556043.1
|
PRMT5
|
protein arginine methyltransferase 5 |
| chr4_-_70653673 | 0.04 |
ENST00000512870.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr2_-_89157161 | 0.04 |
ENST00000390237.2
|
IGKC
|
immunoglobulin kappa constant |
| chr8_+_52730143 | 0.04 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr17_-_57229155 | 0.03 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr3_+_190281229 | 0.03 |
ENST00000453359.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr3_+_108541545 | 0.03 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr2_-_99279928 | 0.03 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr2_-_175712479 | 0.03 |
ENST00000443238.1
|
CHN1
|
chimerin 1 |
| chr4_-_153332886 | 0.03 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr10_-_116286656 | 0.03 |
ENST00000428430.1
ENST00000369266.3 ENST00000392952.3 |
ABLIM1
|
actin binding LIM protein 1 |
| chr2_+_87769459 | 0.03 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr3_-_12587055 | 0.03 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr7_-_151217166 | 0.03 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr17_-_62499334 | 0.03 |
ENST00000579996.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr12_+_6554021 | 0.03 |
ENST00000266557.3
|
CD27
|
CD27 molecule |
| chrX_-_19988382 | 0.03 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr19_-_30199516 | 0.03 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr7_-_112758589 | 0.03 |
ENST00000413744.1
ENST00000439551.1 ENST00000441359.1 |
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr3_-_98619999 | 0.03 |
ENST00000449482.1
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2 |
| chr4_-_186570679 | 0.03 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_-_180707306 | 0.03 |
ENST00000479269.1
|
DNAJC19
|
DnaJ (Hsp40) homolog, subfamily C, member 19 |
| chr11_+_113258495 | 0.03 |
ENST00000303941.3
|
ANKK1
|
ankyrin repeat and kinase domain containing 1 |
| chr16_-_18887627 | 0.03 |
ENST00000563235.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr1_-_89591749 | 0.03 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chrX_+_18725758 | 0.03 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr6_-_66417107 | 0.03 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr7_+_80254279 | 0.03 |
ENST00000436384.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr2_+_145780739 | 0.03 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr3_-_149095652 | 0.03 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr20_+_57464200 | 0.03 |
ENST00000604005.1
|
GNAS
|
GNAS complex locus |
| chr8_+_55528627 | 0.03 |
ENST00000220676.1
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr14_+_29299437 | 0.03 |
ENST00000550827.1
ENST00000548087.1 ENST00000551588.1 ENST00000550122.1 |
CTD-2384A14.1
|
CTD-2384A14.1 |
| chr7_-_5998714 | 0.02 |
ENST00000539903.1
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr10_-_14372870 | 0.02 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr12_-_91573316 | 0.02 |
ENST00000393155.1
|
DCN
|
decorin |
| chr11_-_31531121 | 0.02 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr8_+_92082424 | 0.02 |
ENST00000285420.4
ENST00000404789.3 |
OTUD6B
|
OTU domain containing 6B |
| chr5_+_180682720 | 0.02 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr4_-_120243545 | 0.02 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr18_+_66465475 | 0.02 |
ENST00000581520.1
|
CCDC102B
|
coiled-coil domain containing 102B |
| chr3_-_131221790 | 0.02 |
ENST00000512877.1
ENST00000264995.3 ENST00000511168.1 ENST00000425847.2 |
MRPL3
|
mitochondrial ribosomal protein L3 |
| chr1_-_147142557 | 0.02 |
ENST00000369238.6
|
ACP6
|
acid phosphatase 6, lysophosphatidic |
| chr14_+_52313833 | 0.02 |
ENST00000553560.1
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr1_+_95616933 | 0.02 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr1_+_52682052 | 0.02 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr19_+_20188830 | 0.02 |
ENST00000418063.2
|
ZNF90
|
zinc finger protein 90 |
| chr13_+_33160553 | 0.02 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr4_-_57253587 | 0.02 |
ENST00000513376.1
ENST00000602986.1 ENST00000434343.2 ENST00000451613.1 ENST00000205214.6 ENST00000502617.1 |
AASDH
|
aminoadipate-semialdehyde dehydrogenase |
| chr12_+_128399917 | 0.02 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr2_-_198540751 | 0.02 |
ENST00000429081.1
|
RFTN2
|
raftlin family member 2 |
| chr12_+_128399965 | 0.02 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr14_-_34420259 | 0.02 |
ENST00000250457.3
ENST00000547327.2 |
EGLN3
|
egl-9 family hypoxia-inducible factor 3 |
| chr10_-_29923893 | 0.02 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr3_+_62936098 | 0.02 |
ENST00000475886.1
ENST00000465684.1 ENST00000465262.1 ENST00000468072.1 |
LINC00698
|
long intergenic non-protein coding RNA 698 |
| chr5_+_137722255 | 0.02 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr13_+_24844819 | 0.02 |
ENST00000399949.2
|
SPATA13
|
spermatogenesis associated 13 |
| chr21_-_35014027 | 0.02 |
ENST00000399442.1
ENST00000413017.2 ENST00000445393.1 ENST00000417979.1 ENST00000426935.1 ENST00000381540.3 ENST00000361534.2 ENST00000381554.3 |
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr5_-_41794313 | 0.02 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr3_-_131221771 | 0.02 |
ENST00000510154.1
ENST00000507669.1 |
MRPL3
|
mitochondrial ribosomal protein L3 |
| chr5_+_118668846 | 0.02 |
ENST00000513374.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr16_+_31213206 | 0.01 |
ENST00000561916.2
|
C16orf98
|
chromosome 16 open reading frame 98 |
| chr16_-_23652570 | 0.01 |
ENST00000261584.4
|
PALB2
|
partner and localizer of BRCA2 |
| chr1_+_160709076 | 0.01 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr3_-_100551141 | 0.01 |
ENST00000478235.1
ENST00000471901.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr13_+_24844979 | 0.01 |
ENST00000454083.1
|
SPATA13
|
spermatogenesis associated 13 |
| chr2_+_102953608 | 0.01 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr6_+_53883790 | 0.01 |
ENST00000509997.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr4_-_109541539 | 0.01 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr12_-_74686314 | 0.01 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chrX_+_105937068 | 0.01 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr7_-_65113280 | 0.01 |
ENST00000593865.1
|
AC104057.1
|
Protein LOC100996407 |
| chr3_+_57882024 | 0.01 |
ENST00000494088.1
|
SLMAP
|
sarcolemma associated protein |
| chr1_+_210501589 | 0.01 |
ENST00000413764.2
ENST00000541565.1 |
HHAT
|
hedgehog acyltransferase |
| chr6_+_30848771 | 0.01 |
ENST00000503180.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr16_+_31271274 | 0.01 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr1_+_160370344 | 0.01 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chrX_+_11129388 | 0.01 |
ENST00000321143.4
ENST00000380763.3 ENST00000380762.4 |
HCCS
|
holocytochrome c synthase |
| chr3_-_107099454 | 0.01 |
ENST00000593837.1
ENST00000599431.1 |
RP11-446H18.5
|
RP11-446H18.5 |
| chr7_-_103848405 | 0.01 |
ENST00000447452.2
ENST00000545943.1 ENST00000297431.4 |
ORC5
|
origin recognition complex, subunit 5 |
| chr17_+_48823975 | 0.01 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr6_+_3259148 | 0.01 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr14_+_69865401 | 0.01 |
ENST00000556605.1
ENST00000336643.5 ENST00000031146.4 |
SLC39A9
|
solute carrier family 39, member 9 |
| chr2_+_210517895 | 0.01 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr12_+_48722763 | 0.01 |
ENST00000335017.1
|
H1FNT
|
H1 histone family, member N, testis-specific |
| chr9_-_98268883 | 0.01 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr2_+_87135076 | 0.01 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr8_-_93029865 | 0.01 |
ENST00000422361.2
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr4_-_1670632 | 0.01 |
ENST00000461064.1
|
FAM53A
|
family with sequence similarity 53, member A |
| chr3_+_73110810 | 0.01 |
ENST00000533473.1
|
EBLN2
|
endogenous Bornavirus-like nucleoprotein 2 |
| chr12_-_10978957 | 0.01 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr11_-_108093329 | 0.01 |
ENST00000278612.8
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus |
| chr18_+_28923589 | 0.01 |
ENST00000462981.2
|
DSG1
|
desmoglein 1 |
| chr2_-_166930131 | 0.01 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr1_+_28527070 | 0.01 |
ENST00000596102.1
|
AL353354.2
|
AL353354.2 |
| chr8_-_109260897 | 0.01 |
ENST00000521297.1
ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E
|
eukaryotic translation initiation factor 3, subunit E |
| chr17_-_295730 | 0.01 |
ENST00000329099.4
|
FAM101B
|
family with sequence similarity 101, member B |
| chr2_+_234216454 | 0.01 |
ENST00000447536.1
ENST00000409110.1 |
SAG
|
S-antigen; retina and pineal gland (arrestin) |
| chr2_-_198540719 | 0.01 |
ENST00000295049.4
|
RFTN2
|
raftlin family member 2 |
| chr2_+_179317994 | 0.00 |
ENST00000375129.4
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr21_+_35014706 | 0.00 |
ENST00000399353.1
ENST00000444491.1 ENST00000381318.3 |
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr16_+_23652700 | 0.00 |
ENST00000300087.2
|
DCTN5
|
dynactin 5 (p25) |
| chr10_-_61900762 | 0.00 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr1_+_87012922 | 0.00 |
ENST00000263723.5
|
CLCA4
|
chloride channel accessory 4 |
| chr14_-_22005343 | 0.00 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr1_+_152975488 | 0.00 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr2_+_74648848 | 0.00 |
ENST00000409791.1
ENST00000426787.1 ENST00000348227.4 |
WDR54
|
WD repeat domain 54 |
| chr7_-_14880892 | 0.00 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr3_+_33155444 | 0.00 |
ENST00000320954.6
|
CRTAP
|
cartilage associated protein |
| chr14_+_101297740 | 0.00 |
ENST00000555928.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr21_+_35014783 | 0.00 |
ENST00000381291.4
ENST00000381285.4 ENST00000399367.3 ENST00000399352.1 ENST00000399355.2 ENST00000399349.1 |
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr1_-_85870177 | 0.00 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.0 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.1 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |