Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
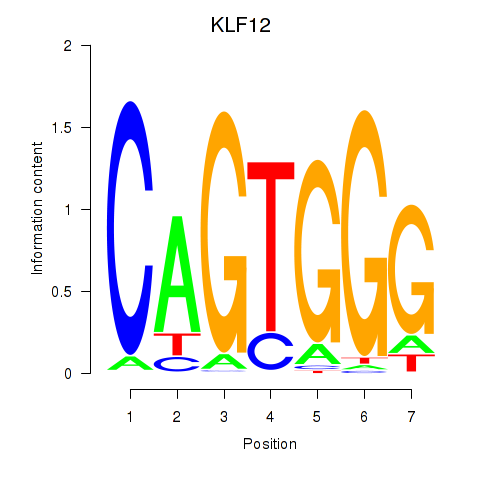
Results for KLF12
Z-value: 1.17
Transcription factors associated with KLF12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF12
|
ENSG00000118922.12 | Kruppel like factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF12 | hg19_v2_chr13_-_74708372_74708409 | -0.25 | 6.3e-01 | Click! |
Activity profile of KLF12 motif
Sorted Z-values of KLF12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_11627248 | 0.99 |
ENST00000510095.1
|
RP11-281P23.2
|
RP11-281P23.2 |
| chr17_-_40264321 | 0.84 |
ENST00000430773.1
ENST00000413196.2 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr1_+_153750622 | 0.73 |
ENST00000532853.1
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr14_+_77425972 | 0.68 |
ENST00000553613.1
|
RP11-7F17.7
|
RP11-7F17.7 |
| chr1_-_47655686 | 0.65 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr2_+_89952792 | 0.59 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr9_+_131084846 | 0.55 |
ENST00000608951.1
|
COQ4
|
coenzyme Q4 |
| chr6_+_27775899 | 0.55 |
ENST00000358739.3
|
HIST1H2AI
|
histone cluster 1, H2ai |
| chr16_-_67978016 | 0.53 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr2_+_90458201 | 0.53 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr9_-_34637806 | 0.50 |
ENST00000477726.1
|
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr19_+_10812108 | 0.49 |
ENST00000250237.5
ENST00000592254.1 |
QTRT1
|
queuine tRNA-ribosyltransferase 1 |
| chr11_-_64856497 | 0.48 |
ENST00000524632.1
ENST00000530719.1 |
AP003068.6
|
transmembrane protein 262 |
| chr16_-_2168079 | 0.47 |
ENST00000488185.2
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr17_-_80797886 | 0.42 |
ENST00000572562.1
|
ZNF750
|
zinc finger protein 750 |
| chr12_-_120907374 | 0.40 |
ENST00000550458.1
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr2_-_85828867 | 0.39 |
ENST00000425160.1
|
TMEM150A
|
transmembrane protein 150A |
| chr14_+_65170820 | 0.39 |
ENST00000555982.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr20_+_44637526 | 0.38 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr12_-_121973974 | 0.37 |
ENST00000538379.1
ENST00000541318.1 ENST00000541511.1 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr1_+_24117627 | 0.33 |
ENST00000400061.1
|
LYPLA2
|
lysophospholipase II |
| chr7_+_70229899 | 0.33 |
ENST00000443672.1
|
AUTS2
|
autism susceptibility candidate 2 |
| chr5_-_180235755 | 0.33 |
ENST00000502678.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr2_+_234104079 | 0.32 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr11_-_57089774 | 0.32 |
ENST00000527207.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr11_+_66360665 | 0.32 |
ENST00000310190.4
|
CCS
|
copper chaperone for superoxide dismutase |
| chr16_-_1843694 | 0.32 |
ENST00000569769.1
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr11_-_102401469 | 0.31 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr10_+_104404644 | 0.31 |
ENST00000462202.2
|
TRIM8
|
tripartite motif containing 8 |
| chr19_+_45312347 | 0.31 |
ENST00000270233.6
ENST00000591520.1 |
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr7_+_66800928 | 0.31 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr17_+_1633755 | 0.31 |
ENST00000545662.1
|
WDR81
|
WD repeat domain 81 |
| chr11_+_65154070 | 0.31 |
ENST00000317568.5
ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8
|
FERM domain containing 8 |
| chr15_-_90039805 | 0.30 |
ENST00000544600.1
ENST00000268122.4 |
RHCG
|
Rh family, C glycoprotein |
| chr6_-_27775694 | 0.30 |
ENST00000377401.2
|
HIST1H2BL
|
histone cluster 1, H2bl |
| chr17_-_47841485 | 0.30 |
ENST00000506156.1
ENST00000240364.2 |
FAM117A
|
family with sequence similarity 117, member A |
| chr17_+_38119216 | 0.29 |
ENST00000301659.4
|
GSDMA
|
gasdermin A |
| chr17_-_72864739 | 0.29 |
ENST00000579893.1
ENST00000544854.1 |
FDXR
|
ferredoxin reductase |
| chr8_-_145086922 | 0.29 |
ENST00000530478.1
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr6_+_31730773 | 0.28 |
ENST00000415669.2
ENST00000425424.1 |
SAPCD1
|
suppressor APC domain containing 1 |
| chr19_+_45174994 | 0.28 |
ENST00000403660.3
|
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr4_-_168155577 | 0.28 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr5_+_137801160 | 0.28 |
ENST00000239938.4
|
EGR1
|
early growth response 1 |
| chr19_-_45909585 | 0.27 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr10_-_77161533 | 0.27 |
ENST00000535216.1
|
ZNF503
|
zinc finger protein 503 |
| chr12_-_54778444 | 0.27 |
ENST00000551771.1
|
ZNF385A
|
zinc finger protein 385A |
| chr5_-_139422654 | 0.26 |
ENST00000289409.4
ENST00000358522.3 ENST00000378238.4 ENST00000289422.7 ENST00000361474.1 ENST00000545385.1 ENST00000394770.1 ENST00000541337.1 |
NRG2
|
neuregulin 2 |
| chr10_-_71906342 | 0.26 |
ENST00000287078.6
ENST00000335494.5 |
TYSND1
|
trypsin domain containing 1 |
| chr11_-_62474803 | 0.26 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr7_+_143013198 | 0.26 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr6_+_31895467 | 0.26 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr2_+_220309379 | 0.25 |
ENST00000451076.1
|
SPEG
|
SPEG complex locus |
| chr17_-_40264692 | 0.25 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr16_+_69140122 | 0.25 |
ENST00000219322.3
|
HAS3
|
hyaluronan synthase 3 |
| chr19_+_40871734 | 0.25 |
ENST00000359274.5
|
PLD3
|
phospholipase D family, member 3 |
| chr4_-_74904398 | 0.24 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr16_+_30671223 | 0.24 |
ENST00000568722.1
|
FBRS
|
fibrosin |
| chr2_-_220435963 | 0.24 |
ENST00000373876.1
ENST00000404537.1 ENST00000603926.1 ENST00000373873.4 ENST00000289656.3 |
OBSL1
|
obscurin-like 1 |
| chr3_+_38032216 | 0.24 |
ENST00000416303.1
|
VILL
|
villin-like |
| chr5_-_121413974 | 0.24 |
ENST00000231004.4
|
LOX
|
lysyl oxidase |
| chr6_+_31895480 | 0.23 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr1_+_239882842 | 0.23 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr19_-_44097188 | 0.23 |
ENST00000594374.1
|
L34079.2
|
L34079.2 |
| chr14_+_102276192 | 0.23 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr19_+_19303720 | 0.22 |
ENST00000392324.4
|
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr17_+_1665253 | 0.22 |
ENST00000254722.4
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr2_+_12858355 | 0.22 |
ENST00000405331.3
|
TRIB2
|
tribbles pseudokinase 2 |
| chr17_-_4889508 | 0.22 |
ENST00000574606.2
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr11_+_61159832 | 0.22 |
ENST00000334888.5
ENST00000398979.3 |
TMEM216
|
transmembrane protein 216 |
| chr3_+_46449049 | 0.22 |
ENST00000357392.4
ENST00000400880.3 ENST00000433848.1 |
CCRL2
|
chemokine (C-C motif) receptor-like 2 |
| chr19_-_56109119 | 0.21 |
ENST00000587678.1
|
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr19_+_49109990 | 0.21 |
ENST00000321762.1
|
SPACA4
|
sperm acrosome associated 4 |
| chr15_+_40531243 | 0.21 |
ENST00000558055.1
ENST00000455577.2 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr1_-_203274418 | 0.21 |
ENST00000457348.1
|
RP11-134P9.1
|
long intergenic non-protein coding RNA 1136 |
| chr19_+_41094612 | 0.21 |
ENST00000595726.1
|
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr17_-_40828969 | 0.21 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr17_-_2117600 | 0.21 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr1_-_119543994 | 0.21 |
ENST00000439394.1
ENST00000449439.1 |
RP4-712E4.1
RP4-712E4.2
|
RP4-712E4.1 RP4-712E4.2 |
| chr12_+_7060508 | 0.20 |
ENST00000541698.1
ENST00000542462.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr11_-_62995986 | 0.20 |
ENST00000403374.2
|
SLC22A25
|
solute carrier family 22, member 25 |
| chr19_-_46000251 | 0.20 |
ENST00000590526.1
ENST00000344680.4 ENST00000245923.4 |
RTN2
|
reticulon 2 |
| chr20_+_34287364 | 0.20 |
ENST00000374072.1
ENST00000397416.1 ENST00000336695.4 |
ROMO1
|
reactive oxygen species modulator 1 |
| chr1_-_114355083 | 0.20 |
ENST00000261441.5
|
RSBN1
|
round spermatid basic protein 1 |
| chr16_-_31519691 | 0.20 |
ENST00000567994.1
ENST00000430477.2 ENST00000570164.1 ENST00000327237.2 |
C16orf58
|
chromosome 16 open reading frame 58 |
| chr14_-_85996332 | 0.19 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr19_+_7600584 | 0.19 |
ENST00000600737.1
|
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr8_+_145691411 | 0.19 |
ENST00000301332.2
|
KIFC2
|
kinesin family member C2 |
| chr16_+_103816 | 0.19 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr11_-_57089671 | 0.19 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr12_+_7060414 | 0.19 |
ENST00000538715.1
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr1_+_109792641 | 0.19 |
ENST00000271332.3
|
CELSR2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr11_-_65374430 | 0.18 |
ENST00000532507.1
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr3_-_61237050 | 0.18 |
ENST00000476844.1
ENST00000488467.1 ENST00000492590.1 ENST00000468189.1 |
FHIT
|
fragile histidine triad |
| chr2_+_220495800 | 0.18 |
ENST00000413743.1
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr19_-_46272106 | 0.18 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5 |
| chr19_+_39214797 | 0.18 |
ENST00000440400.1
|
ACTN4
|
actinin, alpha 4 |
| chr1_+_28918712 | 0.18 |
ENST00000373826.3
|
RAB42
|
RAB42, member RAS oncogene family |
| chr10_-_52645416 | 0.18 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr3_+_52454971 | 0.18 |
ENST00000465863.1
|
PHF7
|
PHD finger protein 7 |
| chr6_+_35773070 | 0.18 |
ENST00000373853.1
ENST00000360215.1 |
LHFPL5
|
lipoma HMGIC fusion partner-like 5 |
| chr16_-_790982 | 0.17 |
ENST00000301694.5
ENST00000251588.2 |
NARFL
|
nuclear prelamin A recognition factor-like |
| chr20_-_36794902 | 0.17 |
ENST00000373403.3
|
TGM2
|
transglutaminase 2 |
| chr15_-_64673630 | 0.17 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr4_-_168155169 | 0.17 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr11_+_45918092 | 0.17 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr6_-_31864977 | 0.17 |
ENST00000395728.3
ENST00000375528.4 |
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr14_+_52313833 | 0.17 |
ENST00000553560.1
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr7_+_2687173 | 0.17 |
ENST00000403167.1
|
TTYH3
|
tweety family member 3 |
| chr11_+_117070037 | 0.17 |
ENST00000392951.4
ENST00000525531.1 ENST00000278968.6 |
TAGLN
|
transgelin |
| chr9_+_131085095 | 0.17 |
ENST00000372875.3
|
COQ4
|
coenzyme Q4 |
| chr22_-_31536480 | 0.17 |
ENST00000215885.3
|
PLA2G3
|
phospholipase A2, group III |
| chr16_+_85646763 | 0.16 |
ENST00000411612.1
ENST00000253458.7 |
GSE1
|
Gse1 coiled-coil protein |
| chr9_+_131084815 | 0.16 |
ENST00000300452.3
ENST00000609948.1 |
COQ4
|
coenzyme Q4 |
| chr3_+_193853927 | 0.16 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr10_-_77161650 | 0.16 |
ENST00000372524.4
|
ZNF503
|
zinc finger protein 503 |
| chr19_-_51412584 | 0.16 |
ENST00000431178.2
|
KLK4
|
kallikrein-related peptidase 4 |
| chr4_-_11430221 | 0.16 |
ENST00000514690.1
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr2_+_172864490 | 0.16 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr11_-_61129335 | 0.16 |
ENST00000545361.1
ENST00000539128.1 ENST00000546151.1 ENST00000447532.2 |
CYB561A3
|
cytochrome b561 family, member A3 |
| chr1_-_76398077 | 0.16 |
ENST00000284142.6
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr7_-_135433534 | 0.16 |
ENST00000338588.3
|
FAM180A
|
family with sequence similarity 180, member A |
| chr1_-_151254362 | 0.16 |
ENST00000447795.2
|
RP11-126K1.2
|
Uncharacterized protein |
| chr10_-_51371321 | 0.15 |
ENST00000602930.1
|
AGAP8
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 8 |
| chr20_+_34287194 | 0.15 |
ENST00000374078.1
ENST00000374077.3 |
ROMO1
|
reactive oxygen species modulator 1 |
| chr19_-_47164386 | 0.15 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr1_+_162336686 | 0.15 |
ENST00000420220.1
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr14_-_74417096 | 0.15 |
ENST00000286544.3
|
FAM161B
|
family with sequence similarity 161, member B |
| chr5_+_145718587 | 0.15 |
ENST00000230732.4
|
POU4F3
|
POU class 4 homeobox 3 |
| chr12_-_117319236 | 0.15 |
ENST00000257572.5
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chrX_+_68048803 | 0.15 |
ENST00000204961.4
|
EFNB1
|
ephrin-B1 |
| chr1_+_1243947 | 0.15 |
ENST00000379031.5
|
PUSL1
|
pseudouridylate synthase-like 1 |
| chr19_+_19303572 | 0.15 |
ENST00000407360.3
ENST00000540981.1 |
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr22_-_24110063 | 0.15 |
ENST00000520222.1
ENST00000401675.3 |
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr16_+_58010339 | 0.15 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr15_-_34394119 | 0.15 |
ENST00000256545.4
|
EMC7
|
ER membrane protein complex subunit 7 |
| chr19_+_46000479 | 0.15 |
ENST00000456399.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr15_+_40531621 | 0.14 |
ENST00000560346.1
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr5_+_139493665 | 0.14 |
ENST00000331327.3
|
PURA
|
purine-rich element binding protein A |
| chr19_+_5914213 | 0.14 |
ENST00000222125.5
ENST00000452990.2 ENST00000588865.1 |
CAPS
|
calcyphosine |
| chr5_+_159343688 | 0.14 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr17_-_39637392 | 0.14 |
ENST00000246639.2
ENST00000393989.1 |
KRT35
|
keratin 35 |
| chr17_+_2240775 | 0.14 |
ENST00000268989.3
ENST00000426855.2 |
SGSM2
|
small G protein signaling modulator 2 |
| chrX_-_48325857 | 0.14 |
ENST00000376875.1
|
SLC38A5
|
solute carrier family 38, member 5 |
| chr17_+_7487146 | 0.14 |
ENST00000396501.4
ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr15_+_74218787 | 0.14 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1 |
| chr17_-_79827808 | 0.14 |
ENST00000580685.1
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr21_-_38445297 | 0.14 |
ENST00000430792.1
ENST00000399103.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr12_+_7060432 | 0.14 |
ENST00000318974.9
ENST00000456013.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr11_+_20620946 | 0.14 |
ENST00000525748.1
|
SLC6A5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr19_+_14063278 | 0.14 |
ENST00000254337.6
|
DCAF15
|
DDB1 and CUL4 associated factor 15 |
| chr17_-_41174367 | 0.14 |
ENST00000587173.1
|
VAT1
|
vesicle amine transport 1 |
| chr16_+_28875126 | 0.14 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr1_+_61330931 | 0.14 |
ENST00000371191.1
|
NFIA
|
nuclear factor I/A |
| chr17_+_27071002 | 0.14 |
ENST00000262395.5
ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4
|
TNF receptor-associated factor 4 |
| chr10_-_103880209 | 0.13 |
ENST00000425280.1
|
LDB1
|
LIM domain binding 1 |
| chr17_+_8191815 | 0.13 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr17_+_7308172 | 0.13 |
ENST00000575301.1
|
NLGN2
|
neuroligin 2 |
| chr16_+_30194916 | 0.13 |
ENST00000570045.1
ENST00000565497.1 ENST00000570244.1 |
CORO1A
|
coronin, actin binding protein, 1A |
| chr3_-_53381539 | 0.13 |
ENST00000606822.1
ENST00000294241.6 ENST00000607628.1 |
DCP1A
|
decapping mRNA 1A |
| chr10_-_32636106 | 0.13 |
ENST00000263062.8
ENST00000319778.6 |
EPC1
|
enhancer of polycomb homolog 1 (Drosophila) |
| chr19_+_4474846 | 0.13 |
ENST00000589486.1
ENST00000592691.1 |
HDGFRP2
|
Hepatoma-derived growth factor-related protein 2 |
| chr9_-_134151915 | 0.13 |
ENST00000372271.3
|
FAM78A
|
family with sequence similarity 78, member A |
| chr5_-_1882858 | 0.13 |
ENST00000511126.1
ENST00000231357.2 |
IRX4
|
iroquois homeobox 4 |
| chr1_-_155177677 | 0.13 |
ENST00000368378.3
ENST00000541990.1 ENST00000457183.2 |
THBS3
|
thrombospondin 3 |
| chr19_+_50381308 | 0.12 |
ENST00000599049.2
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr17_+_17584763 | 0.12 |
ENST00000353383.1
|
RAI1
|
retinoic acid induced 1 |
| chr16_-_30798492 | 0.12 |
ENST00000262525.4
|
ZNF629
|
zinc finger protein 629 |
| chr16_+_811073 | 0.12 |
ENST00000382862.3
ENST00000563651.1 |
MSLN
|
mesothelin |
| chr11_-_118966167 | 0.12 |
ENST00000530167.1
|
H2AFX
|
H2A histone family, member X |
| chr19_+_41092680 | 0.12 |
ENST00000594298.1
ENST00000597396.1 |
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr17_+_57784826 | 0.12 |
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1 |
| chr9_+_139886846 | 0.12 |
ENST00000371620.3
|
C9orf142
|
chromosome 9 open reading frame 142 |
| chr9_-_99064429 | 0.12 |
ENST00000375263.3
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr1_+_16693578 | 0.12 |
ENST00000401088.4
ENST00000471507.1 ENST00000401089.3 ENST00000375590.3 ENST00000492354.1 |
SZRD1
|
SUZ RNA binding domain containing 1 |
| chr22_-_31742218 | 0.12 |
ENST00000266269.5
ENST00000405309.3 ENST00000351933.4 |
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr14_-_24020858 | 0.12 |
ENST00000419474.3
|
ZFHX2
|
zinc finger homeobox 2 |
| chr14_+_65171315 | 0.11 |
ENST00000394691.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr19_+_45174724 | 0.11 |
ENST00000358777.4
|
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr14_+_96343100 | 0.11 |
ENST00000503525.2
|
LINC00617
|
long intergenic non-protein coding RNA 617 |
| chr14_-_74416829 | 0.11 |
ENST00000534936.1
|
FAM161B
|
family with sequence similarity 161, member B |
| chr3_+_52350335 | 0.11 |
ENST00000420323.2
|
DNAH1
|
dynein, axonemal, heavy chain 1 |
| chr3_-_51994694 | 0.11 |
ENST00000395014.2
|
PCBP4
|
poly(rC) binding protein 4 |
| chr10_+_104404218 | 0.11 |
ENST00000302424.7
|
TRIM8
|
tripartite motif containing 8 |
| chr17_-_36906058 | 0.11 |
ENST00000580830.1
|
PCGF2
|
polycomb group ring finger 2 |
| chr16_-_19897455 | 0.11 |
ENST00000568214.1
ENST00000569479.1 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr19_+_50380917 | 0.11 |
ENST00000535102.2
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr17_-_40829026 | 0.11 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr11_-_67120974 | 0.11 |
ENST00000539074.1
ENST00000312419.3 |
POLD4
|
polymerase (DNA-directed), delta 4, accessory subunit |
| chr16_-_2264779 | 0.11 |
ENST00000333503.7
|
PGP
|
phosphoglycolate phosphatase |
| chr3_-_119379427 | 0.11 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr1_-_159832438 | 0.11 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr2_-_97509749 | 0.10 |
ENST00000331001.2
ENST00000318357.4 |
ANKRD23
|
ankyrin repeat domain 23 |
| chr5_-_2751762 | 0.10 |
ENST00000302057.5
ENST00000382611.6 |
IRX2
|
iroquois homeobox 2 |
| chrX_-_74743080 | 0.10 |
ENST00000373367.3
|
ZDHHC15
|
zinc finger, DHHC-type containing 15 |
| chr6_-_134639180 | 0.10 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr2_-_208634287 | 0.10 |
ENST00000295417.3
|
FZD5
|
frizzled family receptor 5 |
| chr2_+_173724771 | 0.10 |
ENST00000538974.1
ENST00000540783.1 |
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_+_36923933 | 0.10 |
ENST00000497382.1
ENST00000404084.1 ENST00000379241.3 ENST00000401530.1 |
VIT
|
vitrin |
| chr16_+_28875268 | 0.10 |
ENST00000395532.4
|
SH2B1
|
SH2B adaptor protein 1 |
| chr2_-_74710078 | 0.10 |
ENST00000290418.4
|
CCDC142
|
coiled-coil domain containing 142 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF12
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.5 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.1 | 0.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.3 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.5 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.1 | 0.3 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.6 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 0.3 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.3 | GO:0050720 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.1 | 0.3 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.2 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.1 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.5 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 0.2 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.6 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.2 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.3 | GO:0010847 | regulation of chromatin assembly(GO:0010847) protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.2 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.3 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.4 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.4 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.1 | GO:1902568 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 1.0 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.0 | 0.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.4 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 0.2 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.2 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0032796 | uropod organization(GO:0032796) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) positive regulation of gastric acid secretion(GO:0060454) response to capsazepine(GO:1901594) |
| 0.0 | 0.1 | GO:0051891 | pancreatic A cell differentiation(GO:0003310) positive regulation of cardioblast differentiation(GO:0051891) |
| 0.0 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.1 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.1 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.4 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:1904252 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) negative regulation of bile acid biosynthetic process(GO:0070858) acinar cell differentiation(GO:0090425) negative regulation of bile acid metabolic process(GO:1904252) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.2 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:1901805 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.2 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.5 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.3 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.2 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.0 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.3 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.1 | 0.5 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.1 | 0.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.3 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.0 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.1 | 0.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.3 | GO:0016730 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.5 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.3 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 0.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.0 | GO:0001002 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |