Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
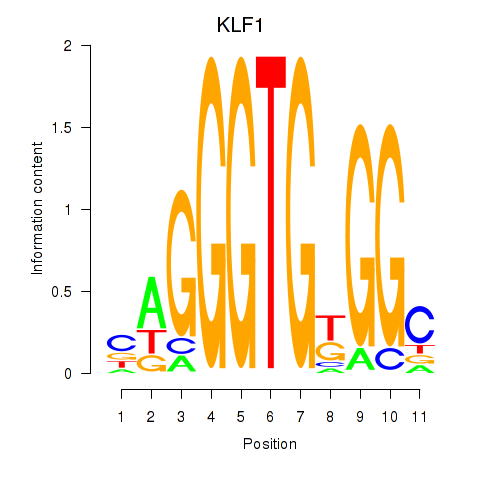
Results for KLF1
Z-value: 1.01
Transcription factors associated with KLF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF1
|
ENSG00000105610.4 | Kruppel like factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF1 | hg19_v2_chr19_-_12997995_12998021 | 0.56 | 2.5e-01 | Click! |
Activity profile of KLF1 motif
Sorted Z-values of KLF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_38171681 | 1.96 |
ENST00000225474.2
ENST00000331769.2 ENST00000394148.3 ENST00000577675.1 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr7_+_22766766 | 1.58 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr17_+_38171614 | 1.31 |
ENST00000583218.1
ENST00000394149.3 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr12_-_49581152 | 0.84 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr5_+_159895275 | 0.76 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr22_-_39639021 | 0.73 |
ENST00000455790.1
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr8_-_42065187 | 0.60 |
ENST00000270189.6
ENST00000352041.3 ENST00000220809.4 |
PLAT
|
plasminogen activator, tissue |
| chr6_+_43968306 | 0.58 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr22_-_30642728 | 0.57 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr16_+_89989687 | 0.56 |
ENST00000315491.7
ENST00000555576.1 ENST00000554336.1 ENST00000553967.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr4_-_82965397 | 0.52 |
ENST00000512716.1
ENST00000514050.1 ENST00000512343.1 ENST00000510780.1 ENST00000508294.1 |
RASGEF1B
RP11-689K5.3
|
RasGEF domain family, member 1B RP11-689K5.3 |
| chr16_-_31161380 | 0.51 |
ENST00000569305.1
ENST00000418068.2 ENST00000268281.4 |
PRSS36
|
protease, serine, 36 |
| chr17_+_38673270 | 0.45 |
ENST00000578280.1
|
RP5-1028K7.2
|
RP5-1028K7.2 |
| chr15_+_45923776 | 0.43 |
ENST00000565227.1
ENST00000563296.1 |
SQRDL
|
sulfide quinone reductase-like (yeast) |
| chr1_+_11866270 | 0.42 |
ENST00000376497.3
ENST00000376487.3 ENST00000376496.3 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr19_+_54960790 | 0.42 |
ENST00000443957.1
|
LENG8
|
leukocyte receptor cluster (LRC) member 8 |
| chr13_-_74708372 | 0.37 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12 |
| chr12_-_56693758 | 0.36 |
ENST00000547298.1
ENST00000551936.1 ENST00000551253.1 ENST00000551473.1 |
CS
|
citrate synthase |
| chr2_-_26205340 | 0.35 |
ENST00000264712.3
|
KIF3C
|
kinesin family member 3C |
| chr2_+_74212073 | 0.34 |
ENST00000441217.1
|
AC073046.25
|
AC073046.25 |
| chr6_+_106546808 | 0.34 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr1_-_26680570 | 0.34 |
ENST00000475866.2
|
AIM1L
|
absent in melanoma 1-like |
| chr17_+_7210921 | 0.34 |
ENST00000573542.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr6_-_3227877 | 0.33 |
ENST00000259818.7
|
TUBB2B
|
tubulin, beta 2B class IIb |
| chr6_-_38607628 | 0.32 |
ENST00000498633.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr22_+_38071615 | 0.32 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr1_+_35225339 | 0.31 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr10_+_104155450 | 0.31 |
ENST00000471698.1
ENST00000189444.6 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr10_+_120967072 | 0.30 |
ENST00000392870.2
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr17_-_79520135 | 0.30 |
ENST00000541246.1
ENST00000544302.1 |
C17orf70
|
chromosome 17 open reading frame 70 |
| chr12_+_120502441 | 0.30 |
ENST00000446727.2
|
CCDC64
|
coiled-coil domain containing 64 |
| chr17_+_7210852 | 0.29 |
ENST00000576930.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr16_+_31483451 | 0.29 |
ENST00000565360.1
ENST00000361773.3 |
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr20_+_44035200 | 0.29 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr2_-_26205550 | 0.28 |
ENST00000405914.1
|
KIF3C
|
kinesin family member 3C |
| chr9_+_74764278 | 0.28 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr17_-_39553844 | 0.28 |
ENST00000251645.2
|
KRT31
|
keratin 31 |
| chr5_-_115910630 | 0.27 |
ENST00000343348.6
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr22_+_31477296 | 0.27 |
ENST00000426927.1
ENST00000440425.1 ENST00000358743.1 ENST00000347557.2 ENST00000333137.7 |
SMTN
|
smoothelin |
| chr16_+_50313426 | 0.26 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chr10_+_17272608 | 0.26 |
ENST00000421459.2
|
VIM
|
vimentin |
| chr15_-_44069513 | 0.26 |
ENST00000433927.1
|
ELL3
|
elongation factor RNA polymerase II-like 3 |
| chr12_+_56498312 | 0.25 |
ENST00000552766.1
|
PA2G4
|
proliferation-associated 2G4, 38kDa |
| chr19_+_39687596 | 0.24 |
ENST00000339852.4
|
NCCRP1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr14_+_32414059 | 0.23 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr6_+_106534192 | 0.23 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr9_+_74764340 | 0.23 |
ENST00000376986.1
ENST00000358399.3 |
GDA
|
guanine deaminase |
| chr20_+_44035847 | 0.22 |
ENST00000372712.2
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr2_-_153573965 | 0.21 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr1_+_11866207 | 0.21 |
ENST00000312413.6
ENST00000346436.6 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr16_+_67312049 | 0.21 |
ENST00000565899.1
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr17_-_39661947 | 0.21 |
ENST00000590425.1
|
KRT13
|
keratin 13 |
| chr16_-_8962200 | 0.20 |
ENST00000562843.1
ENST00000561530.1 ENST00000396593.2 |
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr16_-_31146961 | 0.20 |
ENST00000567531.1
|
PRSS8
|
protease, serine, 8 |
| chr16_-_8962853 | 0.20 |
ENST00000565287.1
ENST00000311052.5 |
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr17_-_27188984 | 0.20 |
ENST00000582320.2
|
MIR144
|
microRNA 451b |
| chr16_-_30125177 | 0.20 |
ENST00000406256.3
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr17_-_1389228 | 0.19 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr1_+_209929446 | 0.19 |
ENST00000479796.1
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr7_-_944631 | 0.19 |
ENST00000453175.2
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr16_-_30134266 | 0.19 |
ENST00000484663.1
ENST00000478356.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr15_+_67390920 | 0.19 |
ENST00000559092.1
ENST00000560175.1 |
SMAD3
|
SMAD family member 3 |
| chr15_-_60690163 | 0.19 |
ENST00000558998.1
ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2
|
annexin A2 |
| chr17_+_1633755 | 0.19 |
ENST00000545662.1
|
WDR81
|
WD repeat domain 81 |
| chr3_+_106959552 | 0.19 |
ENST00000473550.1
|
LINC00883
|
long intergenic non-protein coding RNA 883 |
| chr16_+_30969055 | 0.18 |
ENST00000452917.1
|
SETD1A
|
SET domain containing 1A |
| chr19_-_42916499 | 0.18 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr19_+_41699103 | 0.18 |
ENST00000597754.1
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr3_+_142342240 | 0.18 |
ENST00000497199.1
|
PLS1
|
plastin 1 |
| chr20_-_62710832 | 0.18 |
ENST00000395042.1
|
RGS19
|
regulator of G-protein signaling 19 |
| chr8_-_123793048 | 0.18 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr16_-_58033762 | 0.18 |
ENST00000299237.2
|
ZNF319
|
zinc finger protein 319 |
| chr18_-_5540471 | 0.18 |
ENST00000581833.1
ENST00000544123.1 ENST00000342933.3 ENST00000400111.3 ENST00000585142.1 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr12_-_56694083 | 0.18 |
ENST00000552688.1
ENST00000548041.1 ENST00000551137.1 ENST00000551968.1 ENST00000542324.2 ENST00000546930.1 ENST00000549221.1 ENST00000550159.1 ENST00000550734.1 |
CS
|
citrate synthase |
| chr11_-_64014379 | 0.17 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr22_+_37959647 | 0.17 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr17_-_6917755 | 0.17 |
ENST00000593646.1
|
AC040977.1
|
Uncharacterized protein |
| chr1_+_55505184 | 0.16 |
ENST00000302118.5
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9 |
| chr6_-_31697563 | 0.16 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr20_+_44034804 | 0.16 |
ENST00000357275.2
ENST00000372720.3 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr1_+_43148059 | 0.16 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr1_+_161195835 | 0.16 |
ENST00000545897.1
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like |
| chr5_+_71403061 | 0.16 |
ENST00000512974.1
ENST00000296755.7 |
MAP1B
|
microtubule-associated protein 1B |
| chr18_+_55712915 | 0.16 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr3_+_238456 | 0.16 |
ENST00000427688.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr1_-_24126023 | 0.15 |
ENST00000429356.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr12_-_125398602 | 0.15 |
ENST00000541272.1
ENST00000535131.1 |
UBC
|
ubiquitin C |
| chr1_+_161195781 | 0.15 |
ENST00000367988.3
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like |
| chr17_-_1389419 | 0.15 |
ENST00000575158.1
|
MYO1C
|
myosin IC |
| chr6_+_33589161 | 0.15 |
ENST00000605930.1
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr19_+_36120009 | 0.15 |
ENST00000589871.1
|
RBM42
|
RNA binding motif protein 42 |
| chr9_-_38069208 | 0.15 |
ENST00000377707.3
ENST00000377700.4 |
SHB
|
Src homology 2 domain containing adaptor protein B |
| chr12_-_108154705 | 0.15 |
ENST00000547188.1
|
PRDM4
|
PR domain containing 4 |
| chr5_-_115910091 | 0.15 |
ENST00000257414.8
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr19_+_35629702 | 0.15 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr16_+_58033450 | 0.14 |
ENST00000561743.1
|
USB1
|
U6 snRNA biogenesis 1 |
| chr19_+_8455200 | 0.14 |
ENST00000601897.1
ENST00000594216.1 |
RAB11B
|
RAB11B, member RAS oncogene family |
| chr9_-_35111570 | 0.14 |
ENST00000378561.1
ENST00000603301.1 |
FAM214B
|
family with sequence similarity 214, member B |
| chr1_-_43833628 | 0.14 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr12_+_51632666 | 0.14 |
ENST00000604900.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr5_-_168006591 | 0.14 |
ENST00000239231.6
|
PANK3
|
pantothenate kinase 3 |
| chr9_+_112887772 | 0.14 |
ENST00000259318.7
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr3_-_50383096 | 0.14 |
ENST00000442887.1
ENST00000360165.3 |
ZMYND10
|
zinc finger, MYND-type containing 10 |
| chr11_+_64009072 | 0.14 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr16_+_30075463 | 0.14 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr2_+_85132749 | 0.14 |
ENST00000233143.4
|
TMSB10
|
thymosin beta 10 |
| chr8_-_145018080 | 0.14 |
ENST00000354589.3
|
PLEC
|
plectin |
| chrX_+_47077632 | 0.13 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr19_-_51522955 | 0.13 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr15_-_72521017 | 0.13 |
ENST00000561609.1
|
PKM
|
pyruvate kinase, muscle |
| chr16_+_30075783 | 0.13 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr19_+_7599792 | 0.13 |
ENST00000600942.1
ENST00000593924.1 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr17_+_7210294 | 0.13 |
ENST00000336452.7
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr16_+_56225248 | 0.13 |
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr15_+_44069276 | 0.13 |
ENST00000381359.1
|
SERF2
|
small EDRK-rich factor 2 |
| chr6_-_31697255 | 0.13 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr1_-_153508460 | 0.13 |
ENST00000462776.2
|
S100A6
|
S100 calcium binding protein A6 |
| chr16_+_31483374 | 0.13 |
ENST00000394863.3
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr3_-_186080012 | 0.13 |
ENST00000544847.1
ENST00000265022.3 |
DGKG
|
diacylglycerol kinase, gamma 90kDa |
| chr5_+_89770696 | 0.13 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr20_+_35974532 | 0.13 |
ENST00000373578.2
|
SRC
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr5_+_140501581 | 0.13 |
ENST00000194152.1
|
PCDHB4
|
protocadherin beta 4 |
| chr17_-_46703826 | 0.13 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr2_-_219151984 | 0.13 |
ENST00000444000.1
ENST00000418569.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr9_-_35111420 | 0.13 |
ENST00000378557.1
|
FAM214B
|
family with sequence similarity 214, member B |
| chr14_-_64971288 | 0.13 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chrX_-_153151586 | 0.13 |
ENST00000370060.1
ENST00000370055.1 ENST00000420165.1 |
L1CAM
|
L1 cell adhesion molecule |
| chr4_+_39699664 | 0.13 |
ENST00000261427.5
ENST00000510934.1 ENST00000295963.6 |
UBE2K
|
ubiquitin-conjugating enzyme E2K |
| chr11_-_119252359 | 0.13 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr16_+_30075595 | 0.13 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr5_+_141016508 | 0.13 |
ENST00000444782.1
ENST00000521367.1 ENST00000297164.3 |
RELL2
|
RELT-like 2 |
| chr12_-_58146048 | 0.12 |
ENST00000547281.1
ENST00000546489.1 ENST00000552388.1 ENST00000540325.1 ENST00000312990.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr19_+_46850320 | 0.12 |
ENST00000391919.1
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr14_+_101292445 | 0.12 |
ENST00000429159.2
ENST00000520714.1 ENST00000522771.2 ENST00000424076.3 ENST00000423456.1 ENST00000521404.1 ENST00000556736.1 ENST00000451743.2 ENST00000398518.2 ENST00000554639.1 ENST00000452120.2 ENST00000519709.1 ENST00000412736.2 |
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr19_+_46850251 | 0.12 |
ENST00000012443.4
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr1_-_39395165 | 0.12 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr19_-_10946871 | 0.12 |
ENST00000589638.1
|
TMED1
|
transmembrane emp24 protein transport domain containing 1 |
| chr1_-_113249678 | 0.12 |
ENST00000369633.2
ENST00000425265.2 ENST00000369632.2 ENST00000436685.2 |
RHOC
|
ras homolog family member C |
| chr19_-_51523275 | 0.12 |
ENST00000309958.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr1_-_154155675 | 0.12 |
ENST00000330188.9
ENST00000341485.5 |
TPM3
|
tropomyosin 3 |
| chr2_-_85637459 | 0.12 |
ENST00000409921.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr19_-_18654293 | 0.12 |
ENST00000597547.1
ENST00000222308.4 ENST00000544835.3 ENST00000610101.1 ENST00000597960.3 ENST00000608443.1 |
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr19_-_51523412 | 0.12 |
ENST00000391805.1
ENST00000599077.1 |
KLK10
|
kallikrein-related peptidase 10 |
| chr17_-_66287310 | 0.12 |
ENST00000582867.1
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr15_+_75074410 | 0.12 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr6_-_31763276 | 0.12 |
ENST00000440048.1
|
VARS
|
valyl-tRNA synthetase |
| chr6_+_30850862 | 0.12 |
ENST00000504651.1
ENST00000512694.1 ENST00000515233.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr10_+_94608245 | 0.12 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr11_+_61248583 | 0.12 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr2_+_54684327 | 0.12 |
ENST00000389980.3
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr8_+_120428546 | 0.12 |
ENST00000259526.3
|
NOV
|
nephroblastoma overexpressed |
| chr19_-_10047219 | 0.11 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chr20_-_43743790 | 0.11 |
ENST00000307971.4
ENST00000372789.4 |
WFDC5
|
WAP four-disulfide core domain 5 |
| chr3_-_127542021 | 0.11 |
ENST00000434178.2
|
MGLL
|
monoglyceride lipase |
| chr1_+_203765437 | 0.11 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr6_-_43596899 | 0.11 |
ENST00000307126.5
ENST00000452781.1 |
GTPBP2
|
GTP binding protein 2 |
| chr1_-_159893507 | 0.11 |
ENST00000368096.1
|
TAGLN2
|
transgelin 2 |
| chr7_+_73245193 | 0.11 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr20_+_60878005 | 0.11 |
ENST00000253003.2
|
ADRM1
|
adhesion regulating molecule 1 |
| chr16_-_30596818 | 0.11 |
ENST00000567773.1
|
ZNF785
|
zinc finger protein 785 |
| chr19_+_1104048 | 0.11 |
ENST00000593032.1
ENST00000588919.1 |
GPX4
|
glutathione peroxidase 4 |
| chr1_-_1009683 | 0.11 |
ENST00000453464.2
|
RNF223
|
ring finger protein 223 |
| chr19_-_47349395 | 0.11 |
ENST00000597020.1
|
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr22_-_20138302 | 0.11 |
ENST00000540078.1
ENST00000439765.2 |
AC006547.14
|
uncharacterized protein LOC388849 |
| chr19_+_41698927 | 0.11 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr19_+_47616682 | 0.11 |
ENST00000594526.1
|
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr7_-_1600433 | 0.11 |
ENST00000431208.1
|
TMEM184A
|
transmembrane protein 184A |
| chr1_-_243326612 | 0.11 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr1_+_43148625 | 0.11 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr16_+_30078811 | 0.11 |
ENST00000564688.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr17_-_7518145 | 0.11 |
ENST00000250113.7
ENST00000571597.1 |
FXR2
|
fragile X mental retardation, autosomal homolog 2 |
| chr1_-_113249734 | 0.11 |
ENST00000484054.3
ENST00000369636.2 ENST00000369637.1 ENST00000285735.2 ENST00000369638.2 |
RHOC
|
ras homolog family member C |
| chr17_-_47755338 | 0.11 |
ENST00000508805.1
ENST00000515508.2 ENST00000451526.2 ENST00000507970.1 |
SPOP
|
speckle-type POZ protein |
| chr1_+_209929494 | 0.10 |
ENST00000367026.3
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr17_-_39942322 | 0.10 |
ENST00000449889.1
ENST00000465293.1 |
JUP
|
junction plakoglobin |
| chr12_+_122326630 | 0.10 |
ENST00000541212.1
ENST00000340175.5 |
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr6_+_83903061 | 0.10 |
ENST00000369724.4
ENST00000539997.1 |
RWDD2A
|
RWD domain containing 2A |
| chr17_+_38599693 | 0.10 |
ENST00000542955.1
ENST00000269593.4 |
IGFBP4
|
insulin-like growth factor binding protein 4 |
| chr1_-_94147385 | 0.10 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr11_-_65625014 | 0.10 |
ENST00000534784.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr19_+_36119975 | 0.10 |
ENST00000589559.1
ENST00000360475.4 |
RBM42
|
RNA binding motif protein 42 |
| chr1_-_113249948 | 0.10 |
ENST00000339083.7
ENST00000369642.3 |
RHOC
|
ras homolog family member C |
| chr5_-_172756506 | 0.10 |
ENST00000265087.4
|
STC2
|
stanniocalcin 2 |
| chr17_-_39023462 | 0.10 |
ENST00000251643.4
|
KRT12
|
keratin 12 |
| chrX_+_47078069 | 0.10 |
ENST00000357227.4
ENST00000519758.1 ENST00000520893.1 ENST00000517426.1 |
CDK16
|
cyclin-dependent kinase 16 |
| chr19_+_38664224 | 0.10 |
ENST00000601054.1
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3 |
| chr10_+_104678032 | 0.10 |
ENST00000369878.4
ENST00000369875.3 |
CNNM2
|
cyclin M2 |
| chr6_-_30654977 | 0.10 |
ENST00000399199.3
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18 |
| chr20_-_31124186 | 0.10 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr12_-_125398850 | 0.10 |
ENST00000535859.1
ENST00000546271.1 ENST00000540700.1 ENST00000546120.1 |
UBC
|
ubiquitin C |
| chr9_-_100954910 | 0.10 |
ENST00000375077.4
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr19_+_50919056 | 0.10 |
ENST00000599632.1
|
CTD-2545M3.6
|
CTD-2545M3.6 |
| chr13_+_24553933 | 0.10 |
ENST00000424834.2
ENST00000439928.2 |
SPATA13
RP11-309I15.1
|
spermatogenesis associated 13 RP11-309I15.1 |
| chr4_-_926161 | 0.10 |
ENST00000511163.1
|
GAK
|
cyclin G associated kinase |
| chr3_+_49711391 | 0.10 |
ENST00000296456.5
ENST00000449966.1 |
APEH
|
acylaminoacyl-peptide hydrolase |
| chr4_-_165305086 | 0.10 |
ENST00000507270.1
ENST00000514618.1 ENST00000503008.1 |
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr9_-_35732362 | 0.10 |
ENST00000314888.9
ENST00000540444.1 |
TLN1
|
talin 1 |
| chr1_+_44399466 | 0.10 |
ENST00000498139.2
ENST00000491846.1 |
ARTN
|
artemin |
| chr1_-_6526192 | 0.10 |
ENST00000377782.3
ENST00000351959.5 ENST00000356876.3 |
TNFRSF25
|
tumor necrosis factor receptor superfamily, member 25 |
| chr5_-_141016382 | 0.10 |
ENST00000523088.1
ENST00000305264.3 |
HDAC3
|
histone deacetylase 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.3 | 3.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.2 | 0.6 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.2 | 0.7 | GO:0072255 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.2 | 0.5 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.1 | 0.6 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.7 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.3 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.1 | GO:0061196 | fungiform papilla development(GO:0061196) |
| 0.1 | 0.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.2 | GO:2000656 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.1 | 0.4 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.4 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0044145 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.3 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.2 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.3 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.3 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.6 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.1 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.0 | 0.3 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.1 | GO:1902871 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.2 | GO:2000784 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.2 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.1 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.5 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.3 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.0 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0090649 | rRNA transport(GO:0051029) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.6 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.4 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.2 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.0 | 0.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.2 | 0.6 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 0.5 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.2 | 1.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.2 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.2 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.0 | 0.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.2 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 2.8 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 2.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.0 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |