Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
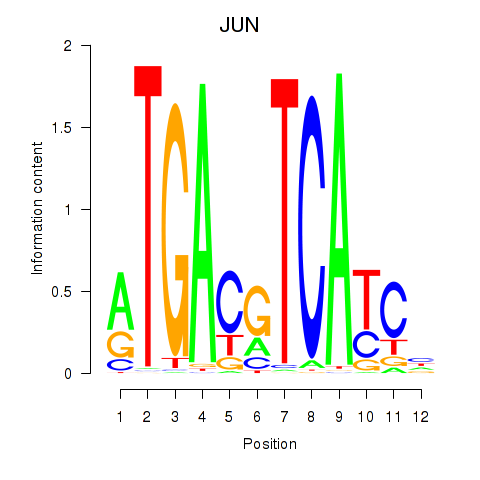
Results for JUN
Z-value: 1.48
Transcription factors associated with JUN
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JUN
|
ENSG00000177606.5 | Jun proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JUN | hg19_v2_chr1_-_59249732_59249785 | 0.58 | 2.3e-01 | Click! |
Activity profile of JUN motif
Sorted Z-values of JUN motif
Network of associatons between targets according to the STRING database.
First level regulatory network of JUN
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.4 | 1.2 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.4 | 1.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.4 | 1.1 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.3 | 0.9 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.3 | 2.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.2 | 0.7 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.2 | 0.6 | GO:0050975 | sensory perception of touch(GO:0050975) maintenance of synapse structure(GO:0099558) presynaptic active zone organization(GO:1990709) |
| 0.2 | 0.6 | GO:0014028 | notochord formation(GO:0014028) axial mesoderm formation(GO:0048320) |
| 0.2 | 1.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 0.7 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.2 | 0.6 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 0.5 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.1 | 0.7 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.5 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.1 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.5 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 0.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 3.1 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 0.6 | GO:1904751 | positive regulation of telomeric DNA binding(GO:1904744) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 0.5 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 0.3 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.1 | 0.3 | GO:1900738 | dense core granule biogenesis(GO:0061110) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of muscle(GO:1901079) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 3.1 | GO:0007567 | parturition(GO:0007567) |
| 0.1 | 0.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.8 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.3 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.1 | 0.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.4 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.1 | 1.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.3 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.4 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.4 | GO:2000724 | positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.1 | 0.2 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.4 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.2 | GO:0043311 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.2 | GO:0061386 | noradrenergic neuron differentiation(GO:0003357) closure of optic fissure(GO:0061386) |
| 0.1 | 0.5 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 1.1 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.1 | 0.2 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.1 | 0.8 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.7 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.3 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.1 | 0.2 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 0.7 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.6 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.2 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.1 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 0.5 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.1 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.1 | 0.2 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.1 | 0.2 | GO:0032826 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.1 | 0.3 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 | 1.0 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 0.2 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.1 | 0.2 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.3 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.7 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.7 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 1.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.2 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.2 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.5 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.2 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.2 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 1.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.3 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.9 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.2 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.3 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.7 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.6 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.5 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.1 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.4 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0035284 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.4 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:1902941 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.5 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 3.0 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) pronephric nephron development(GO:0039019) |
| 0.0 | 0.1 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 1.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.5 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.2 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 0.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.2 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.2 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 1.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.2 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.4 | GO:0030813 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.1 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 1.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.5 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.9 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.1 | GO:2000467 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.2 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.3 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.4 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 1.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 0.7 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.1 | 0.4 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.1 | 0.8 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 0.6 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.3 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.1 | 0.3 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.1 | 0.6 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.6 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.2 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.2 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.1 | 0.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.2 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 3.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 1.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 2.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.2 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.5 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.3 | 1.0 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.3 | 1.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.2 | 0.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 1.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 0.6 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.2 | 0.5 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.2 | 2.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 1.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 1.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 1.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 1.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.4 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 0.8 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 1.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.4 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.3 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.3 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 1.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.6 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.4 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 1.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.2 | GO:0016730 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0086040 | sodium:proton antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086040) |
| 0.0 | 0.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.5 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.4 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 2.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.9 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 1.0 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.2 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.6 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.6 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.2 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 3.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.0 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.0 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 2.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.8 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 2.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 4.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.3 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.9 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 3.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.6 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 2.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 2.8 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |