Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
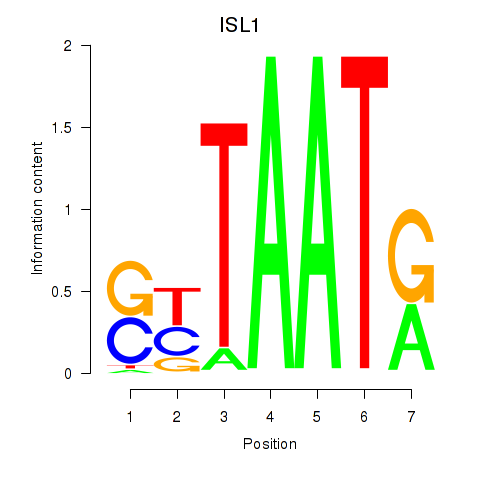
Results for ISL1
Z-value: 0.63
Transcription factors associated with ISL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISL1
|
ENSG00000016082.10 | ISL LIM homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ISL1 | hg19_v2_chr5_+_50679506_50679506 | 0.58 | 2.3e-01 | Click! |
Activity profile of ISL1 motif
Sorted Z-values of ISL1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_24621546 | 0.41 |
ENST00000566108.1
|
CTD-2540M10.1
|
CTD-2540M10.1 |
| chr5_+_137203541 | 0.29 |
ENST00000421631.2
|
MYOT
|
myotilin |
| chr2_-_175711133 | 0.28 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr16_+_47496023 | 0.28 |
ENST00000567200.1
|
PHKB
|
phosphorylase kinase, beta |
| chr17_-_7760457 | 0.26 |
ENST00000576384.1
|
LSMD1
|
LSM domain containing 1 |
| chr17_+_63096903 | 0.25 |
ENST00000582940.1
|
RP11-160O5.1
|
RP11-160O5.1 |
| chr3_-_100551141 | 0.24 |
ENST00000478235.1
ENST00000471901.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr17_-_39123144 | 0.22 |
ENST00000355612.2
|
KRT39
|
keratin 39 |
| chr14_+_21566980 | 0.22 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr11_-_7904464 | 0.21 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr20_-_45530365 | 0.21 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr14_+_62462541 | 0.20 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr2_+_149402009 | 0.20 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr13_-_50020554 | 0.19 |
ENST00000400396.1
|
AL136218.1
|
Sarcoma antigen NY-SAR-79; Uncharacterized protein |
| chr5_+_137203557 | 0.18 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chr12_-_96793142 | 0.18 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr12_-_42631529 | 0.18 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr2_+_47454054 | 0.17 |
ENST00000426892.1
|
AC106869.2
|
AC106869.2 |
| chr20_-_25320367 | 0.16 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chr5_+_137203465 | 0.16 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr3_-_186262166 | 0.15 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr4_+_8201091 | 0.14 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr4_-_186877481 | 0.14 |
ENST00000444781.1
ENST00000432655.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_+_207804278 | 0.14 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr17_+_6918354 | 0.13 |
ENST00000552775.1
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr5_-_94417562 | 0.13 |
ENST00000505465.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr5_-_159766528 | 0.13 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr13_+_78315348 | 0.13 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr14_+_32963433 | 0.13 |
ENST00000554410.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr10_-_17659357 | 0.13 |
ENST00000326961.6
ENST00000361271.3 |
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr5_-_56778635 | 0.13 |
ENST00000423391.1
|
ACTBL2
|
actin, beta-like 2 |
| chr4_-_168155169 | 0.13 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr21_+_17792672 | 0.12 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr14_+_93357749 | 0.12 |
ENST00000557689.1
|
RP11-862G15.1
|
RP11-862G15.1 |
| chr10_-_17659234 | 0.12 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr16_+_3493611 | 0.12 |
ENST00000407558.4
ENST00000572169.1 ENST00000572757.1 ENST00000573593.1 ENST00000570372.1 ENST00000424546.2 ENST00000575733.1 ENST00000573201.1 ENST00000574950.1 ENST00000573580.1 ENST00000608722.1 |
NAA60
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit N-alpha-acetyltransferase 60 |
| chr12_-_57037284 | 0.11 |
ENST00000551570.1
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr15_-_100258029 | 0.11 |
ENST00000378904.2
|
DKFZP779J2370
|
DKFZP779J2370 |
| chrX_+_10031499 | 0.11 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr15_+_76352178 | 0.11 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr11_-_115127611 | 0.11 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr1_+_156024525 | 0.11 |
ENST00000368305.4
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr4_+_71384257 | 0.11 |
ENST00000339336.4
|
AMTN
|
amelotin |
| chr8_-_25281747 | 0.11 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr18_-_19283649 | 0.11 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr12_+_79357815 | 0.10 |
ENST00000547046.1
|
SYT1
|
synaptotagmin I |
| chr2_-_208030295 | 0.10 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr5_+_140593509 | 0.10 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr4_-_77328458 | 0.10 |
ENST00000388914.3
ENST00000434846.2 |
CCDC158
|
coiled-coil domain containing 158 |
| chr8_-_116681686 | 0.10 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr12_+_50144381 | 0.10 |
ENST00000552370.1
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr7_+_138915102 | 0.10 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr1_-_9953295 | 0.10 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr4_-_186877806 | 0.10 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_-_113100088 | 0.10 |
ENST00000374510.4
ENST00000423740.2 ENST00000374511.3 ENST00000374507.4 |
TXNDC8
|
thioredoxin domain containing 8 (spermatozoa) |
| chr6_-_131211534 | 0.10 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr17_-_33885117 | 0.10 |
ENST00000415846.3
|
SLFN14
|
schlafen family member 14 |
| chr5_-_94417339 | 0.10 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr5_+_32710736 | 0.10 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr12_+_9066472 | 0.10 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr9_+_136325089 | 0.10 |
ENST00000291722.7
ENST00000316948.4 ENST00000540581.1 |
CACFD1
|
calcium channel flower domain containing 1 |
| chr19_-_12889226 | 0.09 |
ENST00000589400.1
ENST00000590839.1 ENST00000592079.1 |
HOOK2
|
hook microtubule-tethering protein 2 |
| chr22_-_43355858 | 0.09 |
ENST00000402229.1
ENST00000407585.1 ENST00000453079.1 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr6_-_130031358 | 0.09 |
ENST00000368149.2
|
ARHGAP18
|
Rho GTPase activating protein 18 |
| chr11_-_107729287 | 0.09 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr7_-_44580861 | 0.08 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chr4_-_186734275 | 0.08 |
ENST00000456060.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_+_4861385 | 0.08 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr18_-_52989525 | 0.08 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr10_-_75118611 | 0.08 |
ENST00000355577.3
ENST00000394865.1 ENST00000310715.3 ENST00000401621.2 |
TTC18
|
tetratricopeptide repeat domain 18 |
| chr1_+_156024552 | 0.08 |
ENST00000368304.5
ENST00000368302.3 |
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr18_-_52989217 | 0.07 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr11_-_111649074 | 0.07 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr5_+_135468516 | 0.07 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chr5_+_140248518 | 0.07 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr11_+_7618413 | 0.07 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr2_-_227050079 | 0.07 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr1_-_79472365 | 0.07 |
ENST00000370742.3
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr6_+_90272488 | 0.07 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr2_-_224467093 | 0.07 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr7_+_129932974 | 0.07 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr9_+_131084815 | 0.07 |
ENST00000300452.3
ENST00000609948.1 |
COQ4
|
coenzyme Q4 |
| chr9_-_134585221 | 0.07 |
ENST00000372190.3
ENST00000427994.1 |
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr9_+_131084846 | 0.07 |
ENST00000608951.1
|
COQ4
|
coenzyme Q4 |
| chr4_-_87281196 | 0.06 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr10_+_123923205 | 0.06 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr16_-_23464459 | 0.06 |
ENST00000307149.5
|
COG7
|
component of oligomeric golgi complex 7 |
| chr14_-_21566731 | 0.06 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr2_+_166152283 | 0.06 |
ENST00000375427.2
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr14_-_77495007 | 0.06 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr17_-_15501932 | 0.06 |
ENST00000583965.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr5_-_138211051 | 0.06 |
ENST00000518785.1
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr19_-_22806764 | 0.06 |
ENST00000598042.1
|
AC011516.2
|
AC011516.2 |
| chr1_-_95391315 | 0.06 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chrX_+_49832231 | 0.06 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr6_+_15401075 | 0.06 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr18_-_21977748 | 0.06 |
ENST00000399441.4
ENST00000319481.3 |
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr12_-_94673956 | 0.06 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr8_+_70404996 | 0.06 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr4_+_113970772 | 0.06 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr4_+_169418255 | 0.06 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr2_-_220034745 | 0.06 |
ENST00000455516.2
ENST00000396775.3 ENST00000295738.7 |
SLC23A3
|
solute carrier family 23, member 3 |
| chrX_-_100129128 | 0.06 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr13_+_78315295 | 0.06 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr6_-_116575226 | 0.06 |
ENST00000420283.1
|
TSPYL4
|
TSPY-like 4 |
| chr3_+_121774202 | 0.06 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr5_+_140772381 | 0.06 |
ENST00000398604.2
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr11_-_125351481 | 0.05 |
ENST00000577924.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr7_+_20686946 | 0.05 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr5_-_102455801 | 0.05 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr17_+_1933404 | 0.05 |
ENST00000263083.6
ENST00000571418.1 |
DPH1
|
diphthamide biosynthesis 1 |
| chrX_-_110655306 | 0.05 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr11_-_66496430 | 0.05 |
ENST00000533211.1
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2 |
| chr11_-_107729887 | 0.05 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr18_-_53177984 | 0.05 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr6_+_90272339 | 0.05 |
ENST00000522779.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr19_+_3178736 | 0.05 |
ENST00000246115.3
|
S1PR4
|
sphingosine-1-phosphate receptor 4 |
| chr9_+_128510454 | 0.05 |
ENST00000491787.3
ENST00000447726.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chrX_-_102983417 | 0.05 |
ENST00000372617.4
|
GLRA4
|
glycine receptor, alpha 4 |
| chr7_+_103969104 | 0.05 |
ENST00000424859.1
ENST00000535008.1 ENST00000401970.2 ENST00000543266.1 |
LHFPL3
|
lipoma HMGIC fusion partner-like 3 |
| chr6_+_90272027 | 0.05 |
ENST00000522441.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr1_-_154842741 | 0.05 |
ENST00000271915.4
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr12_-_51418549 | 0.05 |
ENST00000548150.1
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr12_-_11139511 | 0.04 |
ENST00000506868.1
|
TAS2R50
|
taste receptor, type 2, member 50 |
| chr11_-_64660916 | 0.04 |
ENST00000413053.1
|
MIR194-2
|
microRNA 194-2 |
| chr6_+_139135648 | 0.04 |
ENST00000541398.1
|
ECT2L
|
epithelial cell transforming sequence 2 oncogene-like |
| chr8_+_21911054 | 0.04 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr17_-_39023462 | 0.04 |
ENST00000251643.4
|
KRT12
|
keratin 12 |
| chr18_+_55018044 | 0.04 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chrX_-_118986911 | 0.04 |
ENST00000276201.2
ENST00000345865.2 |
UPF3B
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr3_+_57875711 | 0.04 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr17_-_40729681 | 0.04 |
ENST00000590760.1
ENST00000587209.1 ENST00000393795.3 ENST00000253789.5 |
PSMC3IP
|
PSMC3 interacting protein |
| chr1_+_197170592 | 0.04 |
ENST00000535699.1
|
CRB1
|
crumbs homolog 1 (Drosophila) |
| chrX_-_102941596 | 0.04 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr8_-_99955042 | 0.04 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr4_+_104346194 | 0.04 |
ENST00000510200.1
|
RP11-328K4.1
|
RP11-328K4.1 |
| chr14_+_20811766 | 0.04 |
ENST00000250416.5
ENST00000527915.1 |
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chr15_+_59279851 | 0.04 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chrX_-_92928557 | 0.04 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr6_+_31515337 | 0.04 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chrX_-_15619076 | 0.04 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr7_+_99724317 | 0.04 |
ENST00000398075.2
ENST00000421390.1 |
MBLAC1
|
metallo-beta-lactamase domain containing 1 |
| chr5_-_138210977 | 0.04 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr5_+_102200948 | 0.04 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr4_+_159727272 | 0.04 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chrX_-_110513703 | 0.04 |
ENST00000324068.1
|
CAPN6
|
calpain 6 |
| chr1_-_108231101 | 0.04 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr16_+_20775358 | 0.04 |
ENST00000440284.2
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr11_-_46113756 | 0.04 |
ENST00000531959.1
|
PHF21A
|
PHD finger protein 21A |
| chr4_-_68829226 | 0.04 |
ENST00000396188.2
|
TMPRSS11A
|
transmembrane protease, serine 11A |
| chr2_-_217560248 | 0.04 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr4_+_71384300 | 0.04 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr14_+_62585332 | 0.04 |
ENST00000554895.1
|
LINC00643
|
long intergenic non-protein coding RNA 643 |
| chr14_+_37126765 | 0.04 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr4_-_174451370 | 0.04 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr4_-_68829144 | 0.03 |
ENST00000508048.1
|
TMPRSS11A
|
transmembrane protease, serine 11A |
| chr6_+_31707725 | 0.03 |
ENST00000375755.3
ENST00000375742.3 ENST00000375750.3 ENST00000425703.1 ENST00000534153.4 ENST00000375703.3 ENST00000375740.3 |
MSH5
|
mutS homolog 5 |
| chr10_+_114710425 | 0.03 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr9_-_96108696 | 0.03 |
ENST00000375419.1
|
C9orf129
|
chromosome 9 open reading frame 129 |
| chr3_+_130279178 | 0.03 |
ENST00000358511.6
ENST00000453409.2 |
COL6A6
|
collagen, type VI, alpha 6 |
| chr5_-_88180342 | 0.03 |
ENST00000502983.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr18_-_46784778 | 0.03 |
ENST00000582399.1
|
DYM
|
dymeclin |
| chr7_+_16793160 | 0.03 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr17_-_2615031 | 0.03 |
ENST00000576885.1
ENST00000574426.2 |
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr12_-_6484715 | 0.03 |
ENST00000228916.2
|
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chrX_+_65382433 | 0.03 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr11_+_22694123 | 0.03 |
ENST00000534801.1
|
GAS2
|
growth arrest-specific 2 |
| chr16_+_20775024 | 0.03 |
ENST00000289416.5
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr16_-_4664860 | 0.03 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chrX_-_117107680 | 0.03 |
ENST00000447671.2
ENST00000262820.3 |
KLHL13
|
kelch-like family member 13 |
| chr19_-_50143452 | 0.03 |
ENST00000246792.3
|
RRAS
|
related RAS viral (r-ras) oncogene homolog |
| chr12_-_30887948 | 0.03 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr6_+_155537771 | 0.03 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr5_-_41794313 | 0.03 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr2_+_211421262 | 0.03 |
ENST00000233072.5
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr9_+_27109133 | 0.03 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr3_-_121740969 | 0.03 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr17_-_47045949 | 0.03 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr1_+_200011711 | 0.03 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr12_-_52761262 | 0.03 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr6_-_100912785 | 0.03 |
ENST00000369208.3
|
SIM1
|
single-minded family bHLH transcription factor 1 |
| chr7_+_13141097 | 0.03 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr7_-_35734176 | 0.03 |
ENST00000413517.1
ENST00000438224.1 |
HERPUD2
|
HERPUD family member 2 |
| chr2_+_172543919 | 0.03 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr17_-_21156578 | 0.03 |
ENST00000399011.2
ENST00000468196.1 |
C17orf103
|
chromosome 17 open reading frame 103 |
| chr12_+_93096619 | 0.03 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chrX_+_114874727 | 0.03 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr1_+_113217043 | 0.03 |
ENST00000413052.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr17_+_66521936 | 0.03 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chrX_+_65382381 | 0.03 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr11_-_107729504 | 0.03 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr11_+_22688150 | 0.03 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr10_-_18940501 | 0.03 |
ENST00000377304.4
|
NSUN6
|
NOP2/Sun domain family, member 6 |
| chr12_-_10251603 | 0.03 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chrX_-_117119243 | 0.02 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chrX_-_100129320 | 0.02 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
| chr10_-_95241951 | 0.02 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr1_+_113217073 | 0.02 |
ENST00000369645.1
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr11_-_26744908 | 0.02 |
ENST00000533617.1
|
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr6_-_32498046 | 0.02 |
ENST00000374975.3
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ISL1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0045726 | regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.0 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.0 | GO:0003051 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.0 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.0 | 0.0 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.0 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |