Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
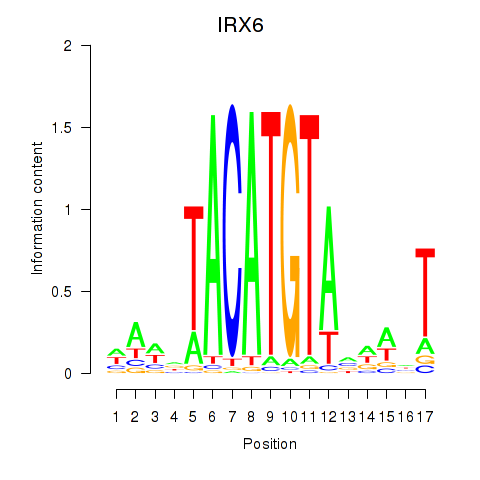
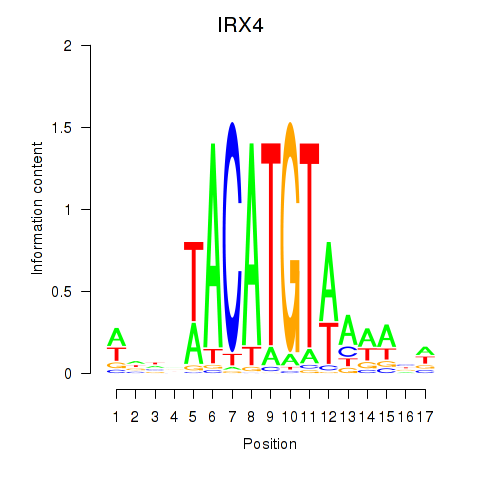
Results for IRX6_IRX4
Z-value: 0.45
Transcription factors associated with IRX6_IRX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX6
|
ENSG00000159387.7 | iroquois homeobox 6 |
|
IRX4
|
ENSG00000113430.5 | iroquois homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX4 | hg19_v2_chr5_-_1882858_1883003 | 0.40 | 4.3e-01 | Click! |
| IRX6 | hg19_v2_chr16_+_55357672_55357672 | 0.27 | 6.0e-01 | Click! |
Activity profile of IRX6_IRX4 motif
Sorted Z-values of IRX6_IRX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_141264597 | 0.35 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr1_+_63989004 | 0.30 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr12_-_123728548 | 0.22 |
ENST00000545406.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr14_-_23623577 | 0.16 |
ENST00000422941.2
ENST00000453702.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr21_-_48024986 | 0.15 |
ENST00000291700.4
ENST00000367071.4 |
S100B
|
S100 calcium binding protein B |
| chr14_-_23624511 | 0.13 |
ENST00000529705.2
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr16_+_777739 | 0.13 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr16_+_777246 | 0.13 |
ENST00000561546.1
ENST00000564545.1 ENST00000389703.3 ENST00000567414.1 ENST00000568141.1 |
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr16_+_776936 | 0.11 |
ENST00000549114.1
ENST00000341413.4 ENST00000562187.1 ENST00000564537.1 |
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr8_-_17555164 | 0.11 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr11_-_35547572 | 0.11 |
ENST00000378880.2
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr1_-_3566590 | 0.11 |
ENST00000424367.1
ENST00000378322.3 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr1_-_201438282 | 0.10 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr1_+_26644441 | 0.09 |
ENST00000374213.2
|
CD52
|
CD52 molecule |
| chr12_+_32655048 | 0.09 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr7_+_6121296 | 0.09 |
ENST00000428901.1
|
AC004895.4
|
AC004895.4 |
| chr12_-_10282836 | 0.08 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr7_-_16844611 | 0.08 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr4_-_104119528 | 0.08 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr1_-_3566627 | 0.08 |
ENST00000419924.2
ENST00000270708.7 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr12_-_10282742 | 0.07 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr1_-_8763278 | 0.07 |
ENST00000468247.1
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr3_-_27498235 | 0.07 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr10_-_115904361 | 0.07 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr4_-_153303658 | 0.07 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr4_-_104119488 | 0.07 |
ENST00000514974.1
|
CENPE
|
centromere protein E, 312kDa |
| chr7_-_55583740 | 0.06 |
ENST00000453256.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr15_+_66585555 | 0.06 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr6_-_33679452 | 0.06 |
ENST00000374231.4
ENST00000607484.1 ENST00000374214.3 |
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr1_+_212475148 | 0.06 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr2_+_160590469 | 0.06 |
ENST00000409591.1
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
| chrX_-_19988382 | 0.05 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr14_+_56127989 | 0.05 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr6_+_149887377 | 0.05 |
ENST00000367419.5
|
GINM1
|
glycoprotein integral membrane 1 |
| chr5_+_36608280 | 0.05 |
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr18_-_28622774 | 0.05 |
ENST00000434452.1
|
DSC3
|
desmocollin 3 |
| chr12_-_76462713 | 0.04 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr12_+_32655110 | 0.04 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr20_+_44330651 | 0.04 |
ENST00000305479.2
|
WFDC13
|
WAP four-disulfide core domain 13 |
| chr15_-_64665911 | 0.04 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr2_+_113829895 | 0.04 |
ENST00000393197.2
|
IL1F10
|
interleukin 1 family, member 10 (theta) |
| chr8_-_101724989 | 0.03 |
ENST00000517403.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr15_+_66585879 | 0.03 |
ENST00000319212.4
|
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr3_+_171561127 | 0.03 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr17_-_29641084 | 0.03 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr3_-_168865522 | 0.03 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr14_+_56127960 | 0.03 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr5_+_66675200 | 0.03 |
ENST00000503106.1
|
RP11-434D9.1
|
RP11-434D9.1 |
| chr15_+_41057818 | 0.03 |
ENST00000558467.1
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr17_-_29641104 | 0.02 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr4_+_144312659 | 0.02 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr11_+_114166536 | 0.02 |
ENST00000299964.3
|
NNMT
|
nicotinamide N-methyltransferase |
| chr5_-_177207634 | 0.02 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr17_-_47723943 | 0.02 |
ENST00000510476.1
ENST00000503676.1 |
SPOP
|
speckle-type POZ protein |
| chr3_-_9994021 | 0.02 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr4_+_113558612 | 0.02 |
ENST00000505034.1
ENST00000324052.6 |
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr3_+_111630451 | 0.02 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr18_-_47376197 | 0.02 |
ENST00000592688.1
|
MYO5B
|
myosin VB |
| chr7_+_871559 | 0.01 |
ENST00000421580.1
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr12_-_88423164 | 0.01 |
ENST00000298699.2
ENST00000550553.1 |
C12orf50
|
chromosome 12 open reading frame 50 |
| chr10_+_90660832 | 0.01 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr1_+_170904612 | 0.01 |
ENST00000367759.4
ENST00000367758.3 |
MROH9
|
maestro heat-like repeat family member 9 |
| chr1_+_53308398 | 0.01 |
ENST00000371528.1
|
ZYG11A
|
zyg-11 family member A, cell cycle regulator |
| chr15_+_81225699 | 0.01 |
ENST00000560027.1
|
KIAA1199
|
KIAA1199 |
| chr4_+_41361616 | 0.00 |
ENST00000513024.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr8_-_67976509 | 0.00 |
ENST00000518747.1
|
COPS5
|
COP9 signalosome subunit 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX6_IRX4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.2 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |