Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
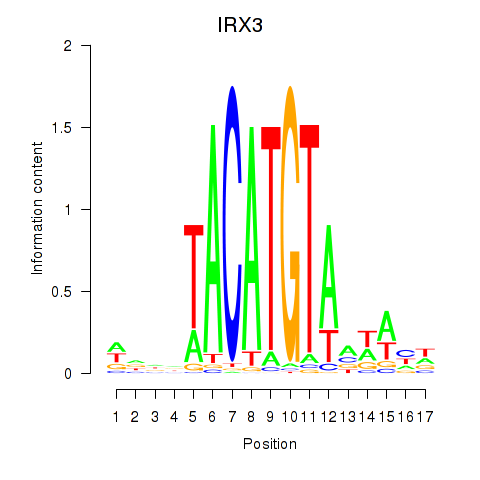
Results for IRX3
Z-value: 0.81
Transcription factors associated with IRX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX3
|
ENSG00000177508.11 | iroquois homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX3 | hg19_v2_chr16_-_54320675_54320715 | 0.32 | 5.4e-01 | Click! |
Activity profile of IRX3 motif
Sorted Z-values of IRX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_58329819 | 0.66 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr15_-_56757329 | 0.56 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr17_+_18380051 | 0.47 |
ENST00000581545.1
ENST00000582333.1 ENST00000328114.6 ENST00000412421.2 ENST00000583322.1 ENST00000584941.1 |
LGALS9C
|
lectin, galactoside-binding, soluble, 9C |
| chr13_+_97928395 | 0.46 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr9_-_4679419 | 0.42 |
ENST00000609131.1
ENST00000607997.1 |
RP11-6J24.6
|
RP11-6J24.6 |
| chr13_+_113030658 | 0.40 |
ENST00000414180.1
ENST00000443541.1 |
SPACA7
|
sperm acrosome associated 7 |
| chr14_+_73563735 | 0.38 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr16_-_25122735 | 0.37 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr22_+_30821732 | 0.36 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr6_+_71104588 | 0.36 |
ENST00000418403.1
|
RP11-462G2.1
|
RP11-462G2.1 |
| chr10_+_9012087 | 0.34 |
ENST00000456526.1
|
RP11-428L9.2
|
RP11-428L9.2 |
| chr1_+_171060018 | 0.34 |
ENST00000367755.4
ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3
|
flavin containing monooxygenase 3 |
| chr4_+_141264597 | 0.33 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr4_-_69434245 | 0.33 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr10_+_90484301 | 0.32 |
ENST00000404190.1
|
LIPK
|
lipase, family member K |
| chr11_+_827248 | 0.29 |
ENST00000527089.1
ENST00000530183.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr2_+_181988620 | 0.28 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr15_+_41057818 | 0.28 |
ENST00000558467.1
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr5_+_169011033 | 0.28 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr2_-_70189397 | 0.27 |
ENST00000320256.4
|
ASPRV1
|
aspartic peptidase, retroviral-like 1 |
| chr2_+_38177575 | 0.27 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr2_-_16804320 | 0.26 |
ENST00000355549.2
|
FAM49A
|
family with sequence similarity 49, member A |
| chr4_+_71091786 | 0.26 |
ENST00000317987.5
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr10_+_82009466 | 0.26 |
ENST00000356374.4
|
AL359195.1
|
Uncharacterized protein; cDNA FLJ46261 fis, clone TESTI4025062 |
| chr5_+_128300810 | 0.26 |
ENST00000262462.4
|
SLC27A6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr3_-_139195350 | 0.26 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr2_+_114163945 | 0.25 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr5_+_81601166 | 0.25 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr4_+_144312659 | 0.24 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr11_+_31531291 | 0.23 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr1_+_87012753 | 0.23 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr1_+_149239529 | 0.23 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr6_+_63921399 | 0.23 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr3_+_14058794 | 0.22 |
ENST00000424053.1
ENST00000528067.1 ENST00000429201.1 |
TPRXL
|
tetra-peptide repeat homeobox-like |
| chr16_+_48657361 | 0.22 |
ENST00000565072.1
|
RP11-42I10.1
|
RP11-42I10.1 |
| chr12_+_55248289 | 0.22 |
ENST00000308796.6
|
MUCL1
|
mucin-like 1 |
| chr3_+_111697843 | 0.21 |
ENST00000534857.1
ENST00000273359.3 ENST00000494817.1 |
ABHD10
|
abhydrolase domain containing 10 |
| chr1_+_87458692 | 0.21 |
ENST00000370548.2
ENST00000356813.4 |
RP5-1052I5.2
HS2ST1
|
Heparan sulfate 2-O-sulfotransferase 1 heparan sulfate 2-O-sulfotransferase 1 |
| chr12_+_21207503 | 0.21 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr6_+_26217159 | 0.21 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr17_+_74729060 | 0.20 |
ENST00000587459.1
|
RP11-318A15.7
|
Uncharacterized protein |
| chr4_-_70725856 | 0.20 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr14_+_39944025 | 0.20 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr4_+_70894130 | 0.19 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr19_-_22379753 | 0.19 |
ENST00000397121.2
|
ZNF676
|
zinc finger protein 676 |
| chr3_+_186383741 | 0.19 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chr9_+_135937365 | 0.19 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr1_+_81001398 | 0.19 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chr18_-_53303123 | 0.18 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr9_+_26746951 | 0.18 |
ENST00000523363.1
|
RP11-18A15.1
|
RP11-18A15.1 |
| chr3_+_177159695 | 0.18 |
ENST00000442937.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr6_-_26108355 | 0.18 |
ENST00000338379.4
|
HIST1H1T
|
histone cluster 1, H1t |
| chr7_-_35013217 | 0.18 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chrX_+_109602039 | 0.18 |
ENST00000520821.1
|
RGAG1
|
retrotransposon gag domain containing 1 |
| chr15_+_49170083 | 0.17 |
ENST00000530028.2
|
EID1
|
EP300 interacting inhibitor of differentiation 1 |
| chr22_+_20877924 | 0.17 |
ENST00000445189.1
|
MED15
|
mediator complex subunit 15 |
| chr5_+_136070614 | 0.17 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr1_-_186365908 | 0.17 |
ENST00000598663.1
|
AL596220.1
|
Uncharacterized protein |
| chr4_+_130017268 | 0.17 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr10_-_90712520 | 0.17 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr9_-_139258159 | 0.17 |
ENST00000371739.3
|
DNLZ
|
DNL-type zinc finger |
| chr20_+_43990576 | 0.16 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr20_-_36156264 | 0.16 |
ENST00000445723.1
ENST00000414080.1 |
BLCAP
|
bladder cancer associated protein |
| chr15_-_100258029 | 0.16 |
ENST00000378904.2
|
DKFZP779J2370
|
DKFZP779J2370 |
| chr18_-_69246186 | 0.16 |
ENST00000568095.1
|
RP11-510D19.1
|
RP11-510D19.1 |
| chr12_-_71182695 | 0.16 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr8_-_50466973 | 0.16 |
ENST00000520800.1
|
RP11-738G5.2
|
Uncharacterized protein |
| chr6_-_26199471 | 0.16 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr12_-_10573149 | 0.15 |
ENST00000381904.2
ENST00000381903.2 ENST00000396439.2 |
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr7_-_55583740 | 0.15 |
ENST00000453256.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr20_-_18477862 | 0.15 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr10_+_35484793 | 0.15 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr18_-_69246167 | 0.15 |
ENST00000566582.1
|
RP11-510D19.1
|
RP11-510D19.1 |
| chr5_-_146833803 | 0.15 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr3_-_100566492 | 0.15 |
ENST00000528490.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr3_+_178865887 | 0.14 |
ENST00000477735.1
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr1_-_179457805 | 0.14 |
ENST00000600581.1
|
AL160286.1
|
Uncharacterized protein |
| chr12_-_100486668 | 0.14 |
ENST00000550544.1
ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L
|
UHRF1 binding protein 1-like |
| chr13_-_38172863 | 0.14 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr16_-_21442874 | 0.14 |
ENST00000534903.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr3_-_48659193 | 0.14 |
ENST00000330862.3
|
TMEM89
|
transmembrane protein 89 |
| chr1_-_62190793 | 0.14 |
ENST00000371177.2
ENST00000606498.1 |
TM2D1
|
TM2 domain containing 1 |
| chr11_-_111782484 | 0.14 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr1_+_1846519 | 0.13 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr19_-_45004556 | 0.13 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr12_+_69742121 | 0.13 |
ENST00000261267.2
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr6_-_26199499 | 0.13 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr13_+_50589390 | 0.13 |
ENST00000360473.4
ENST00000312942.1 |
KCNRG
|
potassium channel regulator |
| chr11_-_63330842 | 0.13 |
ENST00000255695.1
|
HRASLS2
|
HRAS-like suppressor 2 |
| chr11_+_114166536 | 0.13 |
ENST00000299964.3
|
NNMT
|
nicotinamide N-methyltransferase |
| chr7_+_80267973 | 0.13 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr1_-_3566627 | 0.13 |
ENST00000419924.2
ENST00000270708.7 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr17_-_39459103 | 0.13 |
ENST00000391353.1
|
KRTAP29-1
|
keratin associated protein 29-1 |
| chr9_+_130026756 | 0.13 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr11_+_34999328 | 0.12 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr14_-_23623577 | 0.12 |
ENST00000422941.2
ENST00000453702.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr11_+_74303612 | 0.12 |
ENST00000527458.1
ENST00000532497.1 ENST00000530511.1 |
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr21_-_30047095 | 0.12 |
ENST00000452028.1
ENST00000433310.2 |
AF131217.1
|
AF131217.1 |
| chr3_-_48936272 | 0.12 |
ENST00000544097.1
ENST00000430379.1 ENST00000319017.4 |
SLC25A20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr6_+_127587755 | 0.12 |
ENST00000368314.1
ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146
|
ring finger protein 146 |
| chr17_-_29641104 | 0.12 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr19_+_20959098 | 0.12 |
ENST00000360204.5
ENST00000594534.1 |
ZNF66
|
zinc finger protein 66 |
| chr12_-_52867569 | 0.12 |
ENST00000252250.6
|
KRT6C
|
keratin 6C |
| chr8_+_95731904 | 0.12 |
ENST00000522422.1
|
DPY19L4
|
dpy-19-like 4 (C. elegans) |
| chr11_+_27062502 | 0.12 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chrX_-_83757399 | 0.12 |
ENST00000373177.2
ENST00000297977.5 ENST00000506585.2 ENST00000449553.2 |
HDX
|
highly divergent homeobox |
| chr3_+_186692745 | 0.11 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr7_+_80253387 | 0.11 |
ENST00000438020.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr4_-_75695366 | 0.11 |
ENST00000512743.1
|
BTC
|
betacellulin |
| chr1_+_81106951 | 0.11 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr5_+_41904431 | 0.11 |
ENST00000381647.2
|
C5orf51
|
chromosome 5 open reading frame 51 |
| chr1_-_154909329 | 0.11 |
ENST00000368467.3
|
PMVK
|
phosphomevalonate kinase |
| chrX_+_12809463 | 0.11 |
ENST00000380663.3
ENST00000380668.5 ENST00000398491.2 ENST00000489404.1 |
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr15_-_83680325 | 0.11 |
ENST00000508990.2
ENST00000510873.2 ENST00000538348.2 ENST00000451195.3 ENST00000513601.2 ENST00000304177.5 ENST00000565712.1 |
C15orf40
|
chromosome 15 open reading frame 40 |
| chr14_-_23624511 | 0.11 |
ENST00000529705.2
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr4_-_185694872 | 0.11 |
ENST00000505492.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr3_-_178969403 | 0.11 |
ENST00000314235.5
ENST00000392685.2 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr8_-_101724989 | 0.11 |
ENST00000517403.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr19_+_38779778 | 0.10 |
ENST00000590738.1
ENST00000587519.2 ENST00000591889.1 |
SPINT2
CTB-102L5.4
|
serine peptidase inhibitor, Kunitz type, 2 CTB-102L5.4 |
| chr7_-_33080506 | 0.10 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr3_+_8543533 | 0.10 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr11_-_104480019 | 0.10 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr2_+_9983483 | 0.10 |
ENST00000263663.5
|
TAF1B
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa |
| chr1_-_114355083 | 0.10 |
ENST00000261441.5
|
RSBN1
|
round spermatid basic protein 1 |
| chr11_-_82444892 | 0.10 |
ENST00000329203.3
|
FAM181B
|
family with sequence similarity 181, member B |
| chr19_+_56187987 | 0.10 |
ENST00000411543.2
|
EPN1
|
epsin 1 |
| chr4_+_106631966 | 0.10 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr8_+_52730143 | 0.10 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chrX_-_100129128 | 0.10 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr7_+_125078119 | 0.10 |
ENST00000458437.1
ENST00000415896.1 |
RP11-807H17.1
|
RP11-807H17.1 |
| chr4_+_189321881 | 0.10 |
ENST00000512839.1
ENST00000513313.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
| chr2_-_231989808 | 0.10 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr16_-_3493528 | 0.10 |
ENST00000301744.4
|
ZNF597
|
zinc finger protein 597 |
| chr1_+_31883048 | 0.10 |
ENST00000536859.1
|
SERINC2
|
serine incorporator 2 |
| chr19_-_49121054 | 0.10 |
ENST00000546623.1
ENST00000084795.5 |
RPL18
|
ribosomal protein L18 |
| chr6_+_26199737 | 0.10 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr11_+_33037652 | 0.10 |
ENST00000311388.3
|
DEPDC7
|
DEP domain containing 7 |
| chr6_-_41130914 | 0.10 |
ENST00000373113.3
ENST00000338469.3 |
TREM2
|
triggering receptor expressed on myeloid cells 2 |
| chr3_-_58643483 | 0.10 |
ENST00000483787.1
|
FAM3D
|
family with sequence similarity 3, member D |
| chr1_-_201438282 | 0.10 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr17_+_79849872 | 0.09 |
ENST00000584197.1
ENST00000583839.1 |
ANAPC11
|
anaphase promoting complex subunit 11 |
| chr10_-_18944123 | 0.09 |
ENST00000606425.1
|
RP11-139J15.7
|
Uncharacterized protein |
| chr1_+_87012922 | 0.09 |
ENST00000263723.5
|
CLCA4
|
chloride channel accessory 4 |
| chr4_-_69536346 | 0.09 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr1_-_85870177 | 0.09 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr17_-_29641084 | 0.09 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr6_-_135271219 | 0.09 |
ENST00000367847.2
ENST00000367845.2 |
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr2_-_228497888 | 0.09 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr3_+_8543561 | 0.09 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr17_-_42100474 | 0.09 |
ENST00000585950.1
ENST00000592127.1 ENST00000589334.1 |
TMEM101
|
transmembrane protein 101 |
| chr6_-_135271260 | 0.09 |
ENST00000265605.2
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr11_+_86013253 | 0.09 |
ENST00000533986.1
ENST00000278483.3 |
C11orf73
|
chromosome 11 open reading frame 73 |
| chr9_-_3489406 | 0.09 |
ENST00000457373.1
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr19_-_38743878 | 0.09 |
ENST00000587515.1
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr3_+_171561127 | 0.09 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr11_-_111782696 | 0.09 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr14_+_21249200 | 0.09 |
ENST00000304677.2
|
RNASE6
|
ribonuclease, RNase A family, k6 |
| chr14_+_104177607 | 0.09 |
ENST00000429169.1
|
AL049840.1
|
Uncharacterized protein; cDNA FLJ53535 |
| chr2_-_206950996 | 0.09 |
ENST00000414320.1
|
INO80D
|
INO80 complex subunit D |
| chr6_-_144329531 | 0.09 |
ENST00000429150.1
ENST00000392309.1 ENST00000416623.1 ENST00000392307.1 |
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr1_+_215747118 | 0.09 |
ENST00000448333.1
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr16_+_4838412 | 0.09 |
ENST00000589327.1
|
SMIM22
|
small integral membrane protein 22 |
| chr11_+_57531292 | 0.09 |
ENST00000524579.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr11_-_62432641 | 0.09 |
ENST00000528405.1
ENST00000524958.1 ENST00000525675.1 |
RP11-831H9.11
C11orf48
|
Uncharacterized protein chromosome 11 open reading frame 48 |
| chr4_-_108204904 | 0.09 |
ENST00000510463.1
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr12_-_10282742 | 0.09 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr16_-_30032610 | 0.09 |
ENST00000574405.1
|
DOC2A
|
double C2-like domains, alpha |
| chr4_-_70653673 | 0.09 |
ENST00000512870.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr4_-_100212132 | 0.09 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr6_-_18122843 | 0.09 |
ENST00000340650.3
|
NHLRC1
|
NHL repeat containing E3 ubiquitin protein ligase 1 |
| chr1_+_196788887 | 0.09 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr1_+_77333117 | 0.09 |
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr2_+_109204909 | 0.09 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr7_+_127527965 | 0.09 |
ENST00000486037.1
|
SND1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr6_+_159071015 | 0.08 |
ENST00000360448.3
|
SYTL3
|
synaptotagmin-like 3 |
| chr12_+_1738363 | 0.08 |
ENST00000397196.2
|
WNT5B
|
wingless-type MMTV integration site family, member 5B |
| chr5_+_43602750 | 0.08 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr10_-_96829246 | 0.08 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr7_-_6388537 | 0.08 |
ENST00000313324.4
ENST00000530143.1 |
FAM220A
|
family with sequence similarity 220, member A |
| chr11_+_89764274 | 0.08 |
ENST00000448984.1
ENST00000432771.1 |
TRIM49C
|
tripartite motif containing 49C |
| chr8_+_70476088 | 0.08 |
ENST00000525999.1
|
SULF1
|
sulfatase 1 |
| chr2_-_122262593 | 0.08 |
ENST00000418989.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr3_+_8543393 | 0.08 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr19_+_58193337 | 0.08 |
ENST00000601064.1
ENST00000282296.5 ENST00000356715.4 |
ZNF551
|
zinc finger protein 551 |
| chr1_+_114473350 | 0.08 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr8_-_145060593 | 0.08 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr12_+_133757995 | 0.08 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr7_+_77469439 | 0.08 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr12_+_51632638 | 0.08 |
ENST00000549732.2
|
DAZAP2
|
DAZ associated protein 2 |
| chr7_-_112430647 | 0.08 |
ENST00000312814.6
|
TMEM168
|
transmembrane protein 168 |
| chr20_-_23860373 | 0.08 |
ENST00000304710.4
|
CST5
|
cystatin D |
| chr8_+_132952112 | 0.08 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr1_+_63989004 | 0.08 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chrX_+_107288280 | 0.08 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr17_-_46657473 | 0.08 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr4_+_78829479 | 0.08 |
ENST00000504901.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr7_+_99425633 | 0.08 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr12_+_117348742 | 0.08 |
ENST00000309909.5
ENST00000455858.2 |
FBXW8
|
F-box and WD repeat domain containing 8 |
| chr22_-_43355858 | 0.08 |
ENST00000402229.1
ENST00000407585.1 ENST00000453079.1 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr12_-_92536433 | 0.08 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.1 | 0.2 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0002585 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.1 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.3 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.1 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0070839 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) divalent metal ion export(GO:0070839) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.4 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:1901189 | arterial endothelial cell fate commitment(GO:0060844) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.0 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.0 | 0.1 | GO:0036025 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.0 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.0 | 0.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.1 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.2 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.1 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0015227 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 0.1 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.0 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |