Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
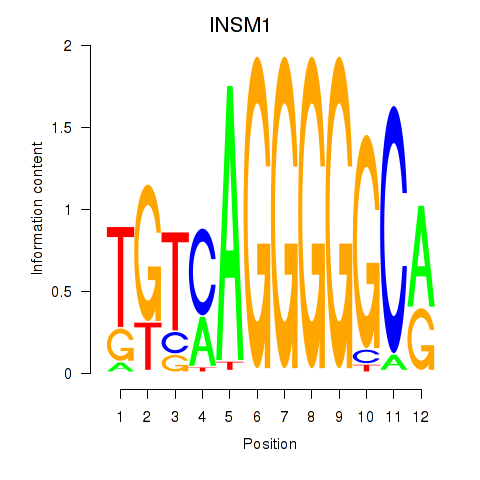
Results for INSM1
Z-value: 0.62
Transcription factors associated with INSM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
INSM1
|
ENSG00000173404.3 | INSM transcriptional repressor 1 |
Activity profile of INSM1 motif
Sorted Z-values of INSM1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_45722727 | 0.62 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr5_-_150466692 | 0.34 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr5_-_150467221 | 0.31 |
ENST00000522226.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr10_+_35416223 | 0.30 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr16_-_30134441 | 0.27 |
ENST00000395200.1
|
MAPK3
|
mitogen-activated protein kinase 3 |
| chr15_-_96590126 | 0.27 |
ENST00000561051.1
|
RP11-4G2.1
|
RP11-4G2.1 |
| chr8_+_99076509 | 0.27 |
ENST00000318528.3
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr16_-_30134524 | 0.21 |
ENST00000395202.1
ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr19_+_48972265 | 0.18 |
ENST00000452733.2
|
CYTH2
|
cytohesin 2 |
| chr15_+_89182178 | 0.18 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr2_-_153573965 | 0.17 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr9_+_136243117 | 0.16 |
ENST00000426926.2
ENST00000371957.3 |
C9orf96
|
chromosome 9 open reading frame 96 |
| chr16_-_19896220 | 0.16 |
ENST00000562469.1
ENST00000300571.2 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr14_+_106938440 | 0.16 |
ENST00000433371.1
ENST00000449670.1 ENST00000334298.3 |
LINC00221
|
long intergenic non-protein coding RNA 221 |
| chr16_+_88704978 | 0.15 |
ENST00000244241.4
|
IL17C
|
interleukin 17C |
| chr3_+_195413160 | 0.15 |
ENST00000599448.1
|
LINC00969
|
long intergenic non-protein coding RNA 969 |
| chr7_+_114055052 | 0.15 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chrX_+_37430822 | 0.15 |
ENST00000378621.3
ENST00000378619.3 |
LANCL3
|
LanC lantibiotic synthetase component C-like 3 (bacterial) |
| chr3_+_14989186 | 0.15 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr2_-_159313214 | 0.14 |
ENST00000409889.1
ENST00000283233.5 ENST00000536771.1 |
CCDC148
|
coiled-coil domain containing 148 |
| chr3_-_39195037 | 0.14 |
ENST00000273153.5
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1 |
| chr11_-_57417367 | 0.13 |
ENST00000534810.1
|
YPEL4
|
yippee-like 4 (Drosophila) |
| chr9_+_137979506 | 0.13 |
ENST00000539529.1
ENST00000392991.4 ENST00000371793.3 |
OLFM1
|
olfactomedin 1 |
| chr1_+_145525015 | 0.12 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr13_-_36871886 | 0.12 |
ENST00000491049.2
ENST00000503173.1 ENST00000239860.6 ENST00000379862.2 ENST00000239859.7 ENST00000379864.2 ENST00000510088.1 ENST00000554962.1 ENST00000511166.1 |
CCDC169
SOHLH2
CCDC169-SOHLH2
|
coiled-coil domain containing 169 spermatogenesis and oogenesis specific basic helix-loop-helix 2 CCDC169-SOHLH2 readthrough |
| chr19_+_48972459 | 0.12 |
ENST00000427476.1
|
CYTH2
|
cytohesin 2 |
| chr19_-_14201776 | 0.12 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr17_-_4889508 | 0.12 |
ENST00000574606.2
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr14_+_31343747 | 0.12 |
ENST00000216361.4
ENST00000396618.3 ENST00000475087.1 |
COCH
|
cochlin |
| chr1_+_53098862 | 0.11 |
ENST00000517870.1
|
FAM159A
|
family with sequence similarity 159, member A |
| chr12_-_92539614 | 0.11 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr1_+_220863187 | 0.11 |
ENST00000294889.5
|
C1orf115
|
chromosome 1 open reading frame 115 |
| chr20_+_35202909 | 0.11 |
ENST00000558530.1
ENST00000558028.1 ENST00000560025.1 |
TGIF2-C20orf24
TGIF2
|
TGIF2-C20orf24 readthrough TGFB-induced factor homeobox 2 |
| chr9_-_140115775 | 0.11 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr9_-_123691439 | 0.11 |
ENST00000540010.1
|
TRAF1
|
TNF receptor-associated factor 1 |
| chr1_+_145524891 | 0.11 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr15_+_90118685 | 0.11 |
ENST00000268138.7
|
TICRR
|
TOPBP1-interacting checkpoint and replication regulator |
| chr8_-_67525524 | 0.10 |
ENST00000517885.1
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr19_+_49977466 | 0.10 |
ENST00000596435.1
ENST00000344019.3 ENST00000597551.1 ENST00000204637.2 ENST00000600429.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr15_+_90118723 | 0.10 |
ENST00000560985.1
|
TICRR
|
TOPBP1-interacting checkpoint and replication regulator |
| chr12_-_51419924 | 0.10 |
ENST00000541174.2
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr19_-_49220084 | 0.10 |
ENST00000595591.1
ENST00000356751.4 ENST00000594582.1 |
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chr13_+_47127293 | 0.10 |
ENST00000311191.6
|
LRCH1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr19_-_48894104 | 0.10 |
ENST00000597017.1
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chrX_-_51812268 | 0.10 |
ENST00000486010.1
ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B
|
melanoma antigen family D, 4B |
| chr5_-_171711061 | 0.10 |
ENST00000393792.2
|
UBTD2
|
ubiquitin domain containing 2 |
| chr20_-_45980621 | 0.10 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr10_+_35415719 | 0.10 |
ENST00000474362.1
ENST00000374721.3 |
CREM
|
cAMP responsive element modulator |
| chr14_+_65879437 | 0.10 |
ENST00000394585.1
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr10_-_104001231 | 0.10 |
ENST00000370002.3
|
PITX3
|
paired-like homeodomain 3 |
| chr6_-_41863098 | 0.09 |
ENST00000373006.1
|
USP49
|
ubiquitin specific peptidase 49 |
| chr17_-_47755338 | 0.09 |
ENST00000508805.1
ENST00000515508.2 ENST00000451526.2 ENST00000507970.1 |
SPOP
|
speckle-type POZ protein |
| chr12_-_51420128 | 0.09 |
ENST00000262051.7
ENST00000547732.1 ENST00000262052.5 ENST00000546488.1 ENST00000550714.1 ENST00000548193.1 ENST00000547579.1 ENST00000546743.1 |
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr15_+_79166065 | 0.09 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr20_+_48807351 | 0.08 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr5_+_154238149 | 0.08 |
ENST00000519430.1
ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr9_-_34637718 | 0.08 |
ENST00000378892.1
ENST00000277010.4 |
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr18_-_34408693 | 0.08 |
ENST00000587382.1
ENST00000589049.1 ENST00000587129.1 |
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chrX_-_71525742 | 0.08 |
ENST00000450875.1
ENST00000417400.1 ENST00000431381.1 ENST00000445983.1 |
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr19_+_38879061 | 0.08 |
ENST00000587013.1
|
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr12_-_6960407 | 0.08 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr9_-_130517522 | 0.08 |
ENST00000373274.3
ENST00000420366.1 |
SH2D3C
|
SH2 domain containing 3C |
| chr19_+_10196781 | 0.08 |
ENST00000253110.11
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr3_+_10857885 | 0.07 |
ENST00000254488.2
ENST00000454147.1 |
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr22_+_20104947 | 0.07 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr22_+_20105012 | 0.07 |
ENST00000331821.3
ENST00000411892.1 |
RANBP1
|
RAN binding protein 1 |
| chr1_+_168148273 | 0.07 |
ENST00000367830.3
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr12_+_58148842 | 0.07 |
ENST00000266643.5
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9 |
| chr16_+_67360712 | 0.07 |
ENST00000569499.2
ENST00000329956.6 ENST00000561948.1 |
LRRC36
|
leucine rich repeat containing 36 |
| chr1_-_36851475 | 0.07 |
ENST00000373129.3
|
STK40
|
serine/threonine kinase 40 |
| chr7_-_128694927 | 0.07 |
ENST00000471166.1
ENST00000265388.5 |
TNPO3
|
transportin 3 |
| chr14_+_31343951 | 0.07 |
ENST00000556908.1
ENST00000555881.1 ENST00000460581.2 |
COCH
|
cochlin |
| chr19_+_49977818 | 0.07 |
ENST00000594009.1
ENST00000595510.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr14_-_85996332 | 0.07 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr2_+_153191706 | 0.07 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr6_+_31126291 | 0.07 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr2_+_232575128 | 0.07 |
ENST00000412128.1
|
PTMA
|
prothymosin, alpha |
| chr1_-_154943002 | 0.07 |
ENST00000606391.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chrX_+_69674943 | 0.07 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr19_+_55795493 | 0.07 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr4_-_147442982 | 0.07 |
ENST00000511374.1
ENST00000264986.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr17_-_1508379 | 0.06 |
ENST00000412517.3
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr16_+_23569021 | 0.06 |
ENST00000567212.1
ENST00000567264.1 |
UBFD1
|
ubiquitin family domain containing 1 |
| chr6_-_31125850 | 0.06 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr15_-_43802769 | 0.06 |
ENST00000263801.3
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr9_+_136243264 | 0.06 |
ENST00000371955.1
|
C9orf96
|
chromosome 9 open reading frame 96 |
| chr19_+_10196981 | 0.06 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr15_+_89182156 | 0.06 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr11_-_57417405 | 0.06 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr1_+_156863470 | 0.06 |
ENST00000338302.3
ENST00000455314.1 ENST00000292357.7 |
PEAR1
|
platelet endothelial aggregation receptor 1 |
| chr18_-_34409116 | 0.06 |
ENST00000334295.4
|
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr17_+_46184911 | 0.06 |
ENST00000580219.1
ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11
|
sorting nexin 11 |
| chr1_-_154943212 | 0.06 |
ENST00000368445.5
ENST00000448116.2 ENST00000368449.4 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr11_+_120110863 | 0.06 |
ENST00000543440.2
|
POU2F3
|
POU class 2 homeobox 3 |
| chr3_+_185303962 | 0.06 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr20_-_45981138 | 0.06 |
ENST00000446994.2
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr6_+_32938692 | 0.06 |
ENST00000443797.2
|
BRD2
|
bromodomain containing 2 |
| chr11_+_58346584 | 0.06 |
ENST00000316059.6
|
ZFP91
|
ZFP91 zinc finger protein |
| chr11_+_8704298 | 0.06 |
ENST00000531978.1
ENST00000524496.1 ENST00000532359.1 ENST00000530022.1 |
RPL27A
|
ribosomal protein L27a |
| chr11_+_10772847 | 0.06 |
ENST00000524523.1
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr1_-_6321035 | 0.06 |
ENST00000377893.2
|
GPR153
|
G protein-coupled receptor 153 |
| chr3_+_185304059 | 0.06 |
ENST00000427465.2
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr16_-_56553882 | 0.06 |
ENST00000568104.1
|
BBS2
|
Bardet-Biedl syndrome 2 |
| chr1_-_36851489 | 0.06 |
ENST00000373130.3
ENST00000373132.3 |
STK40
|
serine/threonine kinase 40 |
| chr11_+_118307179 | 0.06 |
ENST00000534358.1
ENST00000531904.2 ENST00000389506.5 ENST00000354520.4 |
KMT2A
|
lysine (K)-specific methyltransferase 2A |
| chrX_-_153095813 | 0.06 |
ENST00000544474.1
|
PDZD4
|
PDZ domain containing 4 |
| chr17_+_42081914 | 0.06 |
ENST00000293404.3
ENST00000589767.1 |
NAGS
|
N-acetylglutamate synthase |
| chr10_+_35415978 | 0.06 |
ENST00000429130.3
ENST00000469949.2 ENST00000460270.1 |
CREM
|
cAMP responsive element modulator |
| chr10_+_35415851 | 0.06 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chrX_+_51927919 | 0.05 |
ENST00000416960.1
|
MAGED4
|
melanoma antigen family D, 4 |
| chr17_+_45608614 | 0.05 |
ENST00000544660.1
|
NPEPPS
|
aminopeptidase puromycin sensitive |
| chr17_-_32906388 | 0.05 |
ENST00000357754.1
|
C17orf102
|
chromosome 17 open reading frame 102 |
| chr2_-_153573887 | 0.05 |
ENST00000493468.2
ENST00000545856.1 |
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr1_-_154531095 | 0.05 |
ENST00000292211.4
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr14_-_51297360 | 0.05 |
ENST00000496749.1
|
NIN
|
ninein (GSK3B interacting protein) |
| chr17_+_52978185 | 0.05 |
ENST00000572405.1
ENST00000572158.1 ENST00000540336.1 ENST00000572298.1 ENST00000536554.1 ENST00000575333.1 ENST00000570499.1 ENST00000572576.1 |
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr2_+_27435179 | 0.05 |
ENST00000606999.1
ENST00000405489.3 |
ATRAID
|
all-trans retinoic acid-induced differentiation factor |
| chr9_-_37034028 | 0.05 |
ENST00000520281.1
ENST00000446742.1 ENST00000522003.1 ENST00000523145.1 ENST00000414447.1 ENST00000377847.2 ENST00000377853.2 ENST00000377852.2 ENST00000523241.1 ENST00000520154.1 ENST00000358127.4 |
PAX5
|
paired box 5 |
| chr6_+_32938665 | 0.05 |
ENST00000374831.4
ENST00000395289.2 |
BRD2
|
bromodomain containing 2 |
| chr12_-_50101003 | 0.05 |
ENST00000550488.1
|
FMNL3
|
formin-like 3 |
| chr2_-_73511559 | 0.05 |
ENST00000521871.1
|
FBXO41
|
F-box protein 41 |
| chr12_+_3000037 | 0.05 |
ENST00000544943.1
ENST00000448120.2 |
TULP3
|
tubby like protein 3 |
| chr7_-_128695147 | 0.05 |
ENST00000482320.1
ENST00000393245.1 ENST00000471234.1 |
TNPO3
|
transportin 3 |
| chr15_-_77924689 | 0.05 |
ENST00000355300.6
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chrX_-_153095945 | 0.05 |
ENST00000164640.4
|
PDZD4
|
PDZ domain containing 4 |
| chr11_+_124933191 | 0.05 |
ENST00000532000.1
ENST00000308074.4 |
SLC37A2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr1_+_2398876 | 0.05 |
ENST00000449969.1
|
PLCH2
|
phospholipase C, eta 2 |
| chr19_-_39340563 | 0.05 |
ENST00000601813.1
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L |
| chr11_-_46142948 | 0.05 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr12_+_51985001 | 0.04 |
ENST00000354534.6
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr17_-_47755436 | 0.04 |
ENST00000505581.1
ENST00000514121.1 ENST00000393328.2 ENST00000509079.1 ENST00000393331.3 ENST00000347630.2 ENST00000504102.1 |
SPOP
|
speckle-type POZ protein |
| chr12_+_3000073 | 0.04 |
ENST00000397132.2
|
TULP3
|
tubby like protein 3 |
| chr9_-_127533519 | 0.04 |
ENST00000487099.2
ENST00000344523.4 ENST00000373584.3 |
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr17_-_1389228 | 0.04 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr12_+_53774423 | 0.04 |
ENST00000426431.2
|
SP1
|
Sp1 transcription factor |
| chr11_+_10772534 | 0.04 |
ENST00000361367.2
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr3_+_46924829 | 0.04 |
ENST00000313049.5
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr17_-_56606639 | 0.04 |
ENST00000579371.1
|
SEPT4
|
septin 4 |
| chr9_+_115983808 | 0.04 |
ENST00000374210.6
ENST00000374212.4 |
SLC31A1
|
solute carrier family 31 (copper transporter), member 1 |
| chr17_-_56606664 | 0.04 |
ENST00000580844.1
|
SEPT4
|
septin 4 |
| chr17_-_1389419 | 0.04 |
ENST00000575158.1
|
MYO1C
|
myosin IC |
| chr17_-_36762095 | 0.04 |
ENST00000578925.1
ENST00000264659.7 |
SRCIN1
|
SRC kinase signaling inhibitor 1 |
| chr1_-_235116495 | 0.04 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr1_-_160040038 | 0.04 |
ENST00000368089.3
|
KCNJ10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr21_-_46707793 | 0.04 |
ENST00000331343.7
ENST00000349485.5 |
POFUT2
|
protein O-fucosyltransferase 2 |
| chr11_-_64014379 | 0.04 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr11_-_118972575 | 0.04 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr14_+_65879668 | 0.04 |
ENST00000553924.1
ENST00000358307.2 ENST00000557338.1 ENST00000554610.1 |
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr17_-_73389737 | 0.04 |
ENST00000392563.1
|
GRB2
|
growth factor receptor-bound protein 2 |
| chr13_-_52027134 | 0.04 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr22_+_45064593 | 0.04 |
ENST00000432186.1
|
PRR5
|
proline rich 5 (renal) |
| chr18_+_34409069 | 0.04 |
ENST00000543923.1
ENST00000280020.5 ENST00000435985.2 ENST00000592521.1 ENST00000587139.1 |
KIAA1328
|
KIAA1328 |
| chr5_-_141030943 | 0.03 |
ENST00000522783.1
ENST00000519800.1 ENST00000435817.2 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr3_-_15374033 | 0.03 |
ENST00000253688.5
ENST00000383791.3 |
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr12_-_51420108 | 0.03 |
ENST00000547198.1
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chrX_-_153059811 | 0.03 |
ENST00000427365.2
ENST00000444450.1 ENST00000370093.1 |
IDH3G
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr15_+_89181974 | 0.03 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr22_+_38071615 | 0.03 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr17_-_56606705 | 0.03 |
ENST00000317268.3
|
SEPT4
|
septin 4 |
| chr9_-_98269481 | 0.03 |
ENST00000418258.1
ENST00000553011.1 ENST00000551845.1 |
PTCH1
|
patched 1 |
| chr14_-_58332774 | 0.03 |
ENST00000556826.1
|
SLC35F4
|
solute carrier family 35, member F4 |
| chr1_-_99470558 | 0.03 |
ENST00000370188.3
|
LPPR5
|
Lipid phosphate phosphatase-related protein type 5 |
| chr17_-_8868991 | 0.03 |
ENST00000447110.1
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr11_-_57092381 | 0.03 |
ENST00000358252.3
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr18_-_34408802 | 0.03 |
ENST00000590842.1
|
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr17_-_42462688 | 0.03 |
ENST00000377068.3
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr6_-_29595779 | 0.03 |
ENST00000355973.3
ENST00000377012.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr15_-_74045088 | 0.03 |
ENST00000569673.1
|
C15orf59
|
chromosome 15 open reading frame 59 |
| chr11_-_46142615 | 0.03 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chrX_+_153060090 | 0.03 |
ENST00000370086.3
ENST00000370085.3 |
SSR4
|
signal sequence receptor, delta |
| chr17_-_42992856 | 0.03 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr5_+_137688285 | 0.03 |
ENST00000314358.5
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr8_-_101571933 | 0.03 |
ENST00000520311.1
|
ANKRD46
|
ankyrin repeat domain 46 |
| chr6_-_97345689 | 0.03 |
ENST00000316149.7
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
| chr6_+_12749657 | 0.03 |
ENST00000406205.2
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr22_+_44351419 | 0.03 |
ENST00000396202.3
|
SAMM50
|
SAMM50 sorting and assembly machinery component |
| chr1_+_210001309 | 0.03 |
ENST00000491415.2
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish) |
| chr7_-_132262060 | 0.03 |
ENST00000359827.3
|
PLXNA4
|
plexin A4 |
| chr12_+_120105558 | 0.03 |
ENST00000229328.5
ENST00000541640.1 |
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr8_+_42128812 | 0.03 |
ENST00000520810.1
ENST00000416505.2 ENST00000519735.1 ENST00000520835.1 ENST00000379708.3 |
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr12_+_113860160 | 0.03 |
ENST00000553248.1
ENST00000345635.4 ENST00000547802.1 |
SDSL
|
serine dehydratase-like |
| chr9_+_37650945 | 0.03 |
ENST00000377765.3
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr5_+_60933634 | 0.03 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr7_+_86273700 | 0.03 |
ENST00000546348.1
|
GRM3
|
glutamate receptor, metabotropic 3 |
| chr11_+_124932955 | 0.03 |
ENST00000403796.2
|
SLC37A2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr17_-_7297519 | 0.02 |
ENST00000576362.1
ENST00000571078.1 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr5_+_154238042 | 0.02 |
ENST00000519211.1
ENST00000522458.1 ENST00000519903.1 ENST00000521450.1 ENST00000403027.2 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr4_+_26322409 | 0.02 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chrX_-_153059958 | 0.02 |
ENST00000370092.3
ENST00000217901.5 |
IDH3G
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr20_+_44657845 | 0.02 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr2_-_27434635 | 0.02 |
ENST00000401463.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr17_+_37793318 | 0.02 |
ENST00000336308.5
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr9_+_131452239 | 0.02 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr19_+_41284121 | 0.02 |
ENST00000594800.1
ENST00000357052.2 ENST00000602173.1 |
RAB4B
|
RAB4B, member RAS oncogene family |
| chr20_-_30310797 | 0.02 |
ENST00000422920.1
|
BCL2L1
|
BCL2-like 1 |
| chr19_+_10197463 | 0.02 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr8_-_57123815 | 0.02 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr17_+_52978156 | 0.02 |
ENST00000348161.4
|
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr17_-_7297833 | 0.02 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr1_+_10490441 | 0.02 |
ENST00000470413.2
ENST00000309048.3 |
APITD1-CORT
APITD1
|
APITD1-CORT readthrough apoptosis-inducing, TAF9-like domain 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of INSM1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.1 | 0.7 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.2 | GO:0090289 | regulation of osteoclast proliferation(GO:0090289) |
| 0.0 | 0.1 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 0.1 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.2 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.2 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.0 | 0.0 | GO:0010157 | response to chlorate(GO:0010157) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.2 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.1 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.0 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |