Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
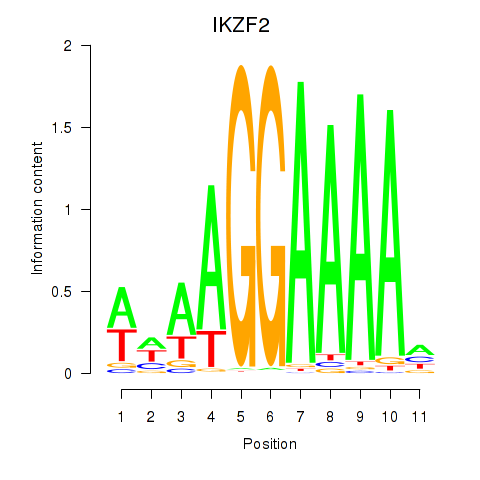
Results for IKZF2
Z-value: 0.57
Transcription factors associated with IKZF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IKZF2
|
ENSG00000030419.12 | IKAROS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | hg19_v2_chr2_-_214016314_214016343 | 0.78 | 6.9e-02 | Click! |
Activity profile of IKZF2 motif
Sorted Z-values of IKZF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_43741594 | 0.34 |
ENST00000536181.1
ENST00000378069.4 |
MAOB
|
monoamine oxidase B |
| chr9_+_92219919 | 0.34 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr5_-_42811986 | 0.31 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr13_+_20268547 | 0.30 |
ENST00000601204.1
|
AL354808.2
|
AL354808.2 |
| chr8_-_72268721 | 0.28 |
ENST00000419131.1
ENST00000388743.2 |
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr14_+_32546145 | 0.28 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr5_-_42812143 | 0.28 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr9_+_67968793 | 0.25 |
ENST00000417488.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr9_-_3469181 | 0.25 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr21_+_17791648 | 0.25 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr12_+_28605426 | 0.23 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr3_+_178865887 | 0.21 |
ENST00000477735.1
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr20_+_52105495 | 0.20 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr13_-_38172863 | 0.20 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chrX_-_45629661 | 0.20 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr10_-_98031310 | 0.18 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr9_-_6015607 | 0.18 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr2_+_114163945 | 0.18 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr4_-_153303658 | 0.17 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr21_+_35107346 | 0.17 |
ENST00000456489.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr14_+_32546274 | 0.17 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr6_-_10435032 | 0.17 |
ENST00000491317.1
ENST00000496285.1 ENST00000479822.1 ENST00000487130.1 |
LINC00518
|
long intergenic non-protein coding RNA 518 |
| chr1_+_40627038 | 0.17 |
ENST00000372771.4
|
RLF
|
rearranged L-myc fusion |
| chr2_+_190722119 | 0.16 |
ENST00000452382.1
|
PMS1
|
PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
| chr1_-_57045228 | 0.16 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr14_-_23479331 | 0.16 |
ENST00000397377.1
ENST00000397379.3 ENST00000406429.2 ENST00000341470.4 ENST00000555998.1 ENST00000397376.2 ENST00000553675.1 ENST00000553931.1 ENST00000555575.1 ENST00000553958.1 ENST00000555098.1 ENST00000556419.1 ENST00000553606.1 ENST00000299088.6 ENST00000554179.1 ENST00000397382.4 |
C14orf93
|
chromosome 14 open reading frame 93 |
| chr12_-_56367101 | 0.15 |
ENST00000549233.2
|
PMEL
|
premelanosome protein |
| chr10_-_116444371 | 0.15 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr1_-_68698197 | 0.15 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr2_+_197504278 | 0.15 |
ENST00000272831.7
ENST00000389175.4 ENST00000472405.2 ENST00000423093.2 |
CCDC150
|
coiled-coil domain containing 150 |
| chr8_-_72268968 | 0.15 |
ENST00000388740.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr18_-_47340297 | 0.14 |
ENST00000586485.1
ENST00000587994.1 ENST00000586100.1 ENST00000285093.10 |
ACAA2
|
acetyl-CoA acyltransferase 2 |
| chr12_-_68845417 | 0.14 |
ENST00000542875.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr6_-_97345689 | 0.14 |
ENST00000316149.7
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
| chr22_+_24199044 | 0.14 |
ENST00000403208.3
ENST00000398356.2 |
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr1_+_95616933 | 0.14 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr10_+_99079008 | 0.14 |
ENST00000371021.3
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas |
| chr8_-_17942432 | 0.14 |
ENST00000381733.4
ENST00000314146.10 |
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| chr4_+_26322409 | 0.13 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr5_+_43602750 | 0.13 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr8_-_133637624 | 0.13 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr15_+_96876340 | 0.13 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_-_101767715 | 0.13 |
ENST00000376840.4
ENST00000409318.1 |
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain) |
| chrX_-_48827976 | 0.13 |
ENST00000218176.3
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr1_+_100111580 | 0.13 |
ENST00000605497.1
|
PALMD
|
palmdelphin |
| chr2_-_174830430 | 0.13 |
ENST00000310015.6
ENST00000455789.2 |
SP3
|
Sp3 transcription factor |
| chr10_-_98031265 | 0.12 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr17_-_6915616 | 0.12 |
ENST00000575889.1
|
AC027763.2
|
Uncharacterized protein |
| chr6_+_32605134 | 0.12 |
ENST00000343139.5
ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr12_-_76461249 | 0.12 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr8_+_97597148 | 0.12 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr2_+_181988620 | 0.12 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr14_-_55493763 | 0.12 |
ENST00000455555.1
ENST00000360586.3 ENST00000421192.1 ENST00000420358.2 |
WDHD1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr4_-_153274078 | 0.12 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr6_+_147830063 | 0.12 |
ENST00000367474.1
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr3_+_177159744 | 0.12 |
ENST00000439009.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr10_+_22614547 | 0.12 |
ENST00000416820.1
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr15_+_69373210 | 0.12 |
ENST00000435479.1
ENST00000559870.1 |
LINC00277
RP11-809H16.5
|
long intergenic non-protein coding RNA 277 RP11-809H16.5 |
| chr10_-_119805558 | 0.12 |
ENST00000369199.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr2_-_214014959 | 0.12 |
ENST00000442445.1
ENST00000457361.1 ENST00000342002.2 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr12_-_6580094 | 0.11 |
ENST00000361716.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr10_-_79398250 | 0.11 |
ENST00000286627.5
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr17_-_25568687 | 0.11 |
ENST00000581944.1
|
RP11-663N22.1
|
RP11-663N22.1 |
| chr17_+_55055466 | 0.11 |
ENST00000262288.3
ENST00000572710.1 ENST00000575395.1 |
SCPEP1
|
serine carboxypeptidase 1 |
| chr4_-_74486109 | 0.11 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr7_+_79765071 | 0.11 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr8_-_72268889 | 0.11 |
ENST00000388742.4
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr21_-_16374688 | 0.11 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr14_-_35591156 | 0.11 |
ENST00000554361.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chrX_-_117119243 | 0.11 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr15_-_90358564 | 0.11 |
ENST00000559874.1
|
ANPEP
|
alanyl (membrane) aminopeptidase |
| chr8_-_6420759 | 0.11 |
ENST00000523120.1
|
ANGPT2
|
angiopoietin 2 |
| chr5_+_94955782 | 0.10 |
ENST00000380007.2
|
GPR150
|
G protein-coupled receptor 150 |
| chr11_+_125464758 | 0.10 |
ENST00000529886.1
|
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr15_-_83837983 | 0.10 |
ENST00000562702.1
|
HDGFRP3
|
Hepatoma-derived growth factor-related protein 3 |
| chr13_+_97874574 | 0.10 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr6_-_11779403 | 0.10 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chrX_+_45364633 | 0.10 |
ENST00000435394.1
ENST00000609127.1 |
RP11-245M24.1
|
RP11-245M24.1 |
| chr1_+_212475148 | 0.10 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr8_+_9183618 | 0.10 |
ENST00000518619.1
|
RP11-115J16.1
|
RP11-115J16.1 |
| chr1_+_117963209 | 0.10 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr1_-_67600639 | 0.10 |
ENST00000544837.1
ENST00000603691.1 |
C1orf141
|
chromosome 1 open reading frame 141 |
| chr1_-_68698222 | 0.10 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chrX_+_80457442 | 0.10 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr2_-_134326009 | 0.10 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr18_-_53089723 | 0.09 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr6_+_32605195 | 0.09 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr1_-_234831899 | 0.09 |
ENST00000442382.1
|
RP4-781K5.6
|
RP4-781K5.6 |
| chr4_-_185776854 | 0.09 |
ENST00000511703.1
|
RP11-701P16.5
|
RP11-701P16.5 |
| chr9_-_88874519 | 0.09 |
ENST00000376001.3
ENST00000339137.3 |
C9orf153
|
chromosome 9 open reading frame 153 |
| chr16_+_28763108 | 0.09 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr8_+_101349823 | 0.09 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr10_-_99030395 | 0.09 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr12_-_100656134 | 0.09 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr21_+_17791838 | 0.09 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr20_-_14318248 | 0.09 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr22_-_29107919 | 0.09 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr3_-_178865747 | 0.09 |
ENST00000435560.1
|
RP11-360P21.2
|
RP11-360P21.2 |
| chr4_+_86525299 | 0.09 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr12_-_118796910 | 0.09 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chrX_+_100663243 | 0.09 |
ENST00000316594.5
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr12_-_118796971 | 0.09 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr17_-_6915646 | 0.08 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
| chr6_-_18265050 | 0.08 |
ENST00000397239.3
|
DEK
|
DEK oncogene |
| chr12_-_80328949 | 0.08 |
ENST00000450142.2
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A |
| chr14_+_23025534 | 0.08 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr1_+_53480598 | 0.08 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr14_+_64970427 | 0.08 |
ENST00000553583.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr5_+_32710736 | 0.08 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr14_+_85994943 | 0.08 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chrX_-_63425561 | 0.08 |
ENST00000374869.3
ENST00000330258.3 |
AMER1
|
APC membrane recruitment protein 1 |
| chr12_-_6602955 | 0.08 |
ENST00000543703.1
|
MRPL51
|
mitochondrial ribosomal protein L51 |
| chr13_-_48669204 | 0.08 |
ENST00000417167.1
|
MED4
|
mediator complex subunit 4 |
| chr8_+_26240414 | 0.08 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr7_-_64451414 | 0.08 |
ENST00000282869.6
|
ZNF117
|
zinc finger protein 117 |
| chr17_-_62499334 | 0.08 |
ENST00000579996.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr14_+_56078695 | 0.08 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr5_+_153418466 | 0.08 |
ENST00000522782.1
ENST00000439768.2 ENST00000436816.1 ENST00000322602.5 ENST00000522177.1 ENST00000520899.1 |
MFAP3
|
microfibrillar-associated protein 3 |
| chr4_-_157892167 | 0.08 |
ENST00000541126.1
|
PDGFC
|
platelet derived growth factor C |
| chr1_+_174933899 | 0.08 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr1_-_151148492 | 0.08 |
ENST00000295314.4
|
TMOD4
|
tropomodulin 4 (muscle) |
| chr1_-_161337662 | 0.08 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr8_-_36636676 | 0.07 |
ENST00000524132.1
ENST00000519451.1 |
RP11-962G15.1
|
RP11-962G15.1 |
| chr9_+_2159850 | 0.07 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_46926048 | 0.07 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr4_-_168155577 | 0.07 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr11_-_75017734 | 0.07 |
ENST00000532525.1
|
ARRB1
|
arrestin, beta 1 |
| chr12_-_110511424 | 0.07 |
ENST00000548191.1
|
C12orf76
|
chromosome 12 open reading frame 76 |
| chr11_+_86502085 | 0.07 |
ENST00000527521.1
|
PRSS23
|
protease, serine, 23 |
| chr10_+_105036909 | 0.07 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha |
| chr16_+_21623958 | 0.07 |
ENST00000568826.1
|
METTL9
|
methyltransferase like 9 |
| chr4_+_95174445 | 0.07 |
ENST00000509418.1
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr8_-_123706338 | 0.07 |
ENST00000521608.1
|
RP11-973F15.1
|
long intergenic non-protein coding RNA 1151 |
| chr3_-_121740969 | 0.07 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr8_-_10987745 | 0.07 |
ENST00000400102.3
|
AF131215.5
|
AF131215.5 |
| chr2_+_46926326 | 0.07 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr10_+_97759848 | 0.07 |
ENST00000424464.1
ENST00000410012.2 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr2_-_101034070 | 0.07 |
ENST00000264249.3
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr14_+_63671105 | 0.07 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr5_+_34685805 | 0.07 |
ENST00000508315.1
|
RAI14
|
retinoic acid induced 14 |
| chr4_-_153456153 | 0.07 |
ENST00000603548.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr11_+_827248 | 0.07 |
ENST00000527089.1
ENST00000530183.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr17_-_57970074 | 0.07 |
ENST00000346141.6
|
TUBD1
|
tubulin, delta 1 |
| chr3_+_178866199 | 0.07 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr6_+_123110302 | 0.07 |
ENST00000368440.4
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chrX_+_123097014 | 0.07 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr19_-_54106751 | 0.07 |
ENST00000600193.1
|
CTB-167G5.5
|
Uncharacterized protein |
| chr1_-_193075180 | 0.07 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr5_-_111091948 | 0.07 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr5_+_32711419 | 0.07 |
ENST00000265074.8
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr1_+_172502244 | 0.07 |
ENST00000610051.1
|
SUCO
|
SUN domain containing ossification factor |
| chr5_+_43603229 | 0.07 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr12_+_50144381 | 0.06 |
ENST00000552370.1
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr13_+_20532848 | 0.06 |
ENST00000382874.2
|
ZMYM2
|
zinc finger, MYM-type 2 |
| chrX_+_52841228 | 0.06 |
ENST00000351072.1
ENST00000425386.1 ENST00000375503.3 |
XAGE5
|
X antigen family, member 5 |
| chr12_-_74686314 | 0.06 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr19_+_18451439 | 0.06 |
ENST00000597431.2
|
PGPEP1
|
pyroglutamyl-peptidase I |
| chrX_+_49832231 | 0.06 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr18_-_21852143 | 0.06 |
ENST00000399443.3
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr12_-_323689 | 0.06 |
ENST00000428720.1
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr10_-_76868931 | 0.06 |
ENST00000372700.3
ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13
|
dual specificity phosphatase 13 |
| chr4_-_74486347 | 0.06 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr6_+_123110465 | 0.06 |
ENST00000539041.1
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr6_-_11779014 | 0.06 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr18_-_24237339 | 0.06 |
ENST00000580191.1
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr12_+_62860581 | 0.06 |
ENST00000393632.2
ENST00000393630.3 ENST00000280379.6 ENST00000546600.1 ENST00000552738.1 ENST00000393629.2 ENST00000552115.1 |
MON2
|
MON2 homolog (S. cerevisiae) |
| chr1_+_104198377 | 0.06 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr6_-_11779174 | 0.06 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr1_-_203151933 | 0.06 |
ENST00000404436.2
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39) |
| chr1_-_48866517 | 0.06 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr7_-_74221288 | 0.06 |
ENST00000451013.2
|
GTF2IRD2
|
GTF2I repeat domain containing 2 |
| chr3_-_46786245 | 0.06 |
ENST00000442359.2
|
PRSS45
|
protease, serine, 45 |
| chr19_+_18451391 | 0.06 |
ENST00000269919.6
ENST00000604499.2 ENST00000595066.1 ENST00000252813.5 |
PGPEP1
|
pyroglutamyl-peptidase I |
| chr2_+_120517717 | 0.06 |
ENST00000420482.1
ENST00000488279.2 |
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr14_-_92506371 | 0.06 |
ENST00000267622.4
|
TRIP11
|
thyroid hormone receptor interactor 11 |
| chr4_-_157892055 | 0.06 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr4_-_113558014 | 0.06 |
ENST00000503172.1
ENST00000505019.1 ENST00000309071.5 |
C4orf21
|
chromosome 4 open reading frame 21 |
| chr1_-_82726817 | 0.06 |
ENST00000420549.1
|
RP11-147G16.1
|
RP11-147G16.1 |
| chr15_-_90358048 | 0.06 |
ENST00000300060.6
ENST00000560137.1 |
ANPEP
|
alanyl (membrane) aminopeptidase |
| chr10_-_112678904 | 0.06 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr3_+_10068095 | 0.06 |
ENST00000287647.3
ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2
|
Fanconi anemia, complementation group D2 |
| chr13_+_20532900 | 0.06 |
ENST00000382871.2
|
ZMYM2
|
zinc finger, MYM-type 2 |
| chr6_-_110964453 | 0.06 |
ENST00000413605.2
|
CDK19
|
cyclin-dependent kinase 19 |
| chr4_-_74486217 | 0.06 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr17_-_56082455 | 0.06 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr1_-_151813033 | 0.05 |
ENST00000454109.1
|
C2CD4D
|
C2 calcium-dependent domain containing 4D |
| chr10_-_38146965 | 0.05 |
ENST00000395873.3
ENST00000357328.4 ENST00000395874.2 |
ZNF248
|
zinc finger protein 248 |
| chr2_+_48796120 | 0.05 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr1_-_197169672 | 0.05 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr10_+_115511434 | 0.05 |
ENST00000369312.4
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr3_-_18466026 | 0.05 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr6_+_26365443 | 0.05 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr1_+_111682058 | 0.05 |
ENST00000545121.1
|
CEPT1
|
choline/ethanolamine phosphotransferase 1 |
| chr4_-_89978299 | 0.05 |
ENST00000511976.1
ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr11_-_19263145 | 0.05 |
ENST00000532666.1
ENST00000527884.1 |
E2F8
|
E2F transcription factor 8 |
| chr11_-_96076334 | 0.05 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr4_+_96012585 | 0.05 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr10_-_24911746 | 0.05 |
ENST00000320481.6
|
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr14_-_57735528 | 0.05 |
ENST00000340918.7
ENST00000413566.2 |
EXOC5
|
exocyst complex component 5 |
| chr5_-_96518907 | 0.05 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IKZF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.3 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.3 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.1 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.5 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.0 | 0.2 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.1 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.1 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.4 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.0 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |