Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
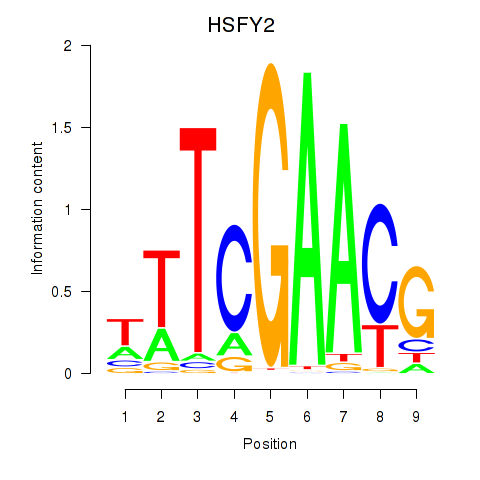
Results for HSFY2
Z-value: 0.92
Transcription factors associated with HSFY2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSFY2
|
ENSG00000169953.11 | heat shock transcription factor Y-linked 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSFY2 | hg19_v2_chrY_-_20935572_20935621 | 0.47 | 3.4e-01 | Click! |
Activity profile of HSFY2 motif
Sorted Z-values of HSFY2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_80263506 | 0.64 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr6_+_63921351 | 0.45 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr1_-_94344686 | 0.41 |
ENST00000528680.1
|
DNTTIP2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr13_+_20268547 | 0.41 |
ENST00000601204.1
|
AL354808.2
|
AL354808.2 |
| chr1_-_89458287 | 0.36 |
ENST00000370485.2
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chr10_+_35416223 | 0.35 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr9_-_25678856 | 0.35 |
ENST00000358022.3
|
TUSC1
|
tumor suppressor candidate 1 |
| chr1_-_89458415 | 0.34 |
ENST00000321792.5
ENST00000370491.3 |
RBMXL1
CCBL2
|
RNA binding motif protein, X-linked-like 1 cysteine conjugate-beta lyase 2 |
| chr3_+_145782358 | 0.34 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chr2_-_169769787 | 0.33 |
ENST00000451987.1
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr7_-_91509972 | 0.33 |
ENST00000425936.1
|
MTERF
|
mitochondrial transcription termination factor |
| chr11_+_125757556 | 0.32 |
ENST00000526028.1
|
HYLS1
|
hydrolethalus syndrome 1 |
| chr12_+_27619743 | 0.32 |
ENST00000298876.4
ENST00000416383.1 |
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr12_-_100486668 | 0.32 |
ENST00000550544.1
ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L
|
UHRF1 binding protein 1-like |
| chr15_+_45694567 | 0.32 |
ENST00000559860.1
|
SPATA5L1
|
spermatogenesis associated 5-like 1 |
| chr7_-_35840198 | 0.32 |
ENST00000412856.1
ENST00000437235.3 ENST00000424194.1 |
AC007551.3
|
AC007551.3 |
| chr5_+_68485363 | 0.31 |
ENST00000283006.2
ENST00000515001.1 |
CENPH
|
centromere protein H |
| chr4_+_146402346 | 0.31 |
ENST00000514778.1
ENST00000507594.1 |
SMAD1
|
SMAD family member 1 |
| chr14_+_21525981 | 0.30 |
ENST00000308227.2
|
RNASE8
|
ribonuclease, RNase A family, 8 |
| chr6_-_73935163 | 0.30 |
ENST00000370388.3
|
KHDC1L
|
KH homology domain containing 1-like |
| chr1_-_197115818 | 0.29 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr15_+_96904487 | 0.29 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr15_-_35280426 | 0.28 |
ENST00000559564.1
ENST00000356321.4 |
ZNF770
|
zinc finger protein 770 |
| chr14_+_39944025 | 0.28 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr16_-_18923035 | 0.27 |
ENST00000563836.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr8_+_120428546 | 0.27 |
ENST00000259526.3
|
NOV
|
nephroblastoma overexpressed |
| chr3_-_186262166 | 0.27 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr3_-_165555200 | 0.27 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr7_+_99425633 | 0.26 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr14_+_39583427 | 0.26 |
ENST00000308317.6
ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2
|
gem (nuclear organelle) associated protein 2 |
| chr1_+_234509413 | 0.26 |
ENST00000366613.1
ENST00000366612.1 |
COA6
|
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr3_-_129375556 | 0.26 |
ENST00000510323.1
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr19_-_43421989 | 0.25 |
ENST00000292125.2
ENST00000187910.2 |
PSG6
|
pregnancy specific beta-1-glycoprotein 6 |
| chr6_-_107077347 | 0.25 |
ENST00000369063.3
ENST00000539449.1 |
RTN4IP1
|
reticulon 4 interacting protein 1 |
| chr2_+_74056147 | 0.25 |
ENST00000394070.2
ENST00000536064.1 |
STAMBP
|
STAM binding protein |
| chr6_-_72129806 | 0.24 |
ENST00000413945.1
ENST00000602878.1 ENST00000436803.1 ENST00000421704.1 ENST00000441570.1 |
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr8_-_61429315 | 0.24 |
ENST00000530725.1
ENST00000532232.1 |
RP11-163N6.2
|
RP11-163N6.2 |
| chr10_-_104866395 | 0.24 |
ENST00000458345.1
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr10_-_90611566 | 0.23 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr14_+_22600515 | 0.23 |
ENST00000390456.3
|
TRAV8-7
|
T cell receptor alpha variable 8-7 (non-functional) |
| chr2_+_181988620 | 0.23 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr6_-_26033796 | 0.23 |
ENST00000259791.2
|
HIST1H2AB
|
histone cluster 1, H2ab |
| chr5_+_68485433 | 0.22 |
ENST00000502689.1
|
CENPH
|
centromere protein H |
| chr2_+_74056066 | 0.22 |
ENST00000339566.3
ENST00000409707.1 ENST00000452725.1 ENST00000432295.2 ENST00000424659.1 ENST00000394073.1 |
STAMBP
|
STAM binding protein |
| chrX_+_155110956 | 0.22 |
ENST00000286448.6
ENST00000262640.6 ENST00000460621.1 |
VAMP7
|
vesicle-associated membrane protein 7 |
| chr3_-_20227619 | 0.22 |
ENST00000425061.1
ENST00000443724.1 ENST00000421451.1 ENST00000452020.1 ENST00000417364.1 ENST00000306698.2 ENST00000419233.2 ENST00000263753.4 ENST00000383774.1 ENST00000437051.1 ENST00000412868.1 ENST00000429446.3 ENST00000442720.1 |
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chr9_-_79009414 | 0.22 |
ENST00000376736.1
|
RFK
|
riboflavin kinase |
| chr9_+_116343192 | 0.22 |
ENST00000471324.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr18_+_57567180 | 0.22 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr5_+_667759 | 0.22 |
ENST00000594226.1
|
AC026740.1
|
Uncharacterized protein |
| chr19_-_33182616 | 0.22 |
ENST00000592431.1
|
CTD-2538C1.2
|
CTD-2538C1.2 |
| chr10_-_105110890 | 0.21 |
ENST00000369847.3
|
PCGF6
|
polycomb group ring finger 6 |
| chr18_+_39535152 | 0.21 |
ENST00000262039.4
ENST00000398870.3 ENST00000586545.1 |
PIK3C3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr20_+_62697564 | 0.21 |
ENST00000458442.1
|
TCEA2
|
transcription elongation factor A (SII), 2 |
| chr4_+_130017268 | 0.21 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr12_-_111142729 | 0.20 |
ENST00000546713.1
|
HVCN1
|
hydrogen voltage-gated channel 1 |
| chr3_+_186383741 | 0.20 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chr13_-_30160925 | 0.20 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr2_-_27435634 | 0.20 |
ENST00000430186.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chrX_+_100353153 | 0.20 |
ENST00000423383.1
ENST00000218507.5 ENST00000403304.2 ENST00000435570.1 |
CENPI
|
centromere protein I |
| chr7_+_79764104 | 0.20 |
ENST00000351004.3
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr1_-_38157877 | 0.19 |
ENST00000477060.1
ENST00000491981.1 ENST00000488137.1 |
C1orf109
|
chromosome 1 open reading frame 109 |
| chr9_+_131901710 | 0.19 |
ENST00000524946.2
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr1_-_53163992 | 0.19 |
ENST00000371538.3
|
SELRC1
|
cytochrome c oxidase assembly factor 7 |
| chr3_-_155011483 | 0.19 |
ENST00000489090.1
|
RP11-451G4.2
|
RP11-451G4.2 |
| chr1_-_226496898 | 0.19 |
ENST00000481685.1
|
LIN9
|
lin-9 homolog (C. elegans) |
| chr1_-_222763240 | 0.18 |
ENST00000352967.4
ENST00000391882.1 ENST00000543857.1 |
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr15_-_57025759 | 0.18 |
ENST00000267807.7
|
ZNF280D
|
zinc finger protein 280D |
| chr5_+_68860949 | 0.18 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr17_+_68047418 | 0.18 |
ENST00000586373.1
ENST00000588782.1 |
LINC01028
|
long intergenic non-protein coding RNA 1028 |
| chr3_+_3168600 | 0.18 |
ENST00000251607.6
ENST00000339437.6 ENST00000280591.6 ENST00000420393.1 |
TRNT1
|
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr6_+_26538566 | 0.18 |
ENST00000377575.2
|
HMGN4
|
high mobility group nucleosomal binding domain 4 |
| chr12_+_66217911 | 0.18 |
ENST00000403681.2
|
HMGA2
|
high mobility group AT-hook 2 |
| chr7_+_23637763 | 0.18 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr5_-_37371278 | 0.17 |
ENST00000231498.3
|
NUP155
|
nucleoporin 155kDa |
| chr10_-_95462265 | 0.17 |
ENST00000536233.1
ENST00000359204.4 ENST00000371430.2 ENST00000394100.2 |
FRA10AC1
|
fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 |
| chr18_+_39535239 | 0.17 |
ENST00000585528.1
|
PIK3C3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr12_+_117013656 | 0.17 |
ENST00000556529.1
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr18_-_61329118 | 0.17 |
ENST00000332821.8
ENST00000283752.5 |
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr11_+_17281900 | 0.17 |
ENST00000530527.1
|
NUCB2
|
nucleobindin 2 |
| chrX_+_21857717 | 0.17 |
ENST00000379484.5
|
MBTPS2
|
membrane-bound transcription factor peptidase, site 2 |
| chr6_+_160211481 | 0.16 |
ENST00000367034.4
|
MRPL18
|
mitochondrial ribosomal protein L18 |
| chrX_+_52841228 | 0.16 |
ENST00000351072.1
ENST00000425386.1 ENST00000375503.3 |
XAGE5
|
X antigen family, member 5 |
| chr1_-_52456352 | 0.16 |
ENST00000371655.3
|
RAB3B
|
RAB3B, member RAS oncogene family |
| chr19_-_1876156 | 0.16 |
ENST00000565797.1
|
CTB-31O20.2
|
CTB-31O20.2 |
| chr10_+_105881779 | 0.16 |
ENST00000369729.3
|
SFR1
|
SWI5-dependent recombination repair 1 |
| chr8_+_144120648 | 0.16 |
ENST00000395172.1
|
C8orf31
|
chromosome 8 open reading frame 31 |
| chr8_+_48920960 | 0.16 |
ENST00000523111.2
ENST00000523432.1 ENST00000521346.1 ENST00000517630.1 |
UBE2V2
|
ubiquitin-conjugating enzyme E2 variant 2 |
| chr6_+_53976285 | 0.16 |
ENST00000514433.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr15_+_45694523 | 0.16 |
ENST00000305560.6
|
SPATA5L1
|
spermatogenesis associated 5-like 1 |
| chr4_-_68995571 | 0.16 |
ENST00000356291.2
|
TMPRSS11F
|
transmembrane protease, serine 11F |
| chr3_+_138067314 | 0.16 |
ENST00000423968.2
|
MRAS
|
muscle RAS oncogene homolog |
| chr1_-_110950255 | 0.15 |
ENST00000483260.1
ENST00000474861.2 ENST00000602318.1 |
LAMTOR5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr8_-_120259055 | 0.15 |
ENST00000522828.1
ENST00000523307.1 ENST00000524129.1 ENST00000521048.1 ENST00000522187.1 |
RP11-4K16.2
|
RP11-4K16.2 |
| chr19_-_52489923 | 0.15 |
ENST00000593596.1
ENST00000243644.4 ENST00000594929.1 ENST00000601430.1 |
ZNF350
|
zinc finger protein 350 |
| chr1_+_93646238 | 0.15 |
ENST00000448243.1
ENST00000370276.1 |
CCDC18
|
coiled-coil domain containing 18 |
| chr4_-_68749699 | 0.15 |
ENST00000545541.1
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr7_+_105172532 | 0.15 |
ENST00000257700.2
|
RINT1
|
RAD50 interactor 1 |
| chr11_-_92930556 | 0.15 |
ENST00000529184.1
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr14_+_104394770 | 0.15 |
ENST00000409874.4
ENST00000339063.5 |
TDRD9
|
tudor domain containing 9 |
| chr11_+_102980126 | 0.15 |
ENST00000375735.2
|
DYNC2H1
|
dynein, cytoplasmic 2, heavy chain 1 |
| chr14_-_39639523 | 0.15 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chr12_-_8088773 | 0.15 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr17_-_46178741 | 0.15 |
ENST00000581003.1
ENST00000225603.4 |
CBX1
|
chromobox homolog 1 |
| chr19_+_36705504 | 0.15 |
ENST00000456324.1
|
ZNF146
|
zinc finger protein 146 |
| chr6_-_26285737 | 0.14 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr3_-_64431146 | 0.14 |
ENST00000569824.1
ENST00000485770.1 |
PRICKLE2
RP11-14D22.2
|
prickle homolog 2 (Drosophila) RP11-14D22.2 |
| chr13_-_34250861 | 0.14 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr3_+_25831567 | 0.14 |
ENST00000280701.3
ENST00000420173.2 |
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr3_-_148804275 | 0.14 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chr12_-_21910775 | 0.14 |
ENST00000539782.1
|
LDHB
|
lactate dehydrogenase B |
| chr8_+_59323823 | 0.14 |
ENST00000399598.2
|
UBXN2B
|
UBX domain protein 2B |
| chr10_-_36812323 | 0.14 |
ENST00000543053.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr20_-_52612705 | 0.14 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr10_+_17272608 | 0.13 |
ENST00000421459.2
|
VIM
|
vimentin |
| chr11_+_86748998 | 0.13 |
ENST00000525018.1
ENST00000355734.4 |
TMEM135
|
transmembrane protein 135 |
| chr7_-_93520191 | 0.13 |
ENST00000545378.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr3_+_101280677 | 0.13 |
ENST00000309922.6
ENST00000495642.1 |
TRMT10C
|
tRNA methyltransferase 10 homolog C (S. cerevisiae) |
| chr3_+_122044084 | 0.13 |
ENST00000264474.3
ENST00000479204.1 |
CSTA
|
cystatin A (stefin A) |
| chr10_-_91403625 | 0.13 |
ENST00000322191.6
ENST00000342512.3 ENST00000371774.2 |
PANK1
|
pantothenate kinase 1 |
| chr12_+_12878829 | 0.13 |
ENST00000326765.6
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr17_-_8113542 | 0.13 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr11_-_61101247 | 0.13 |
ENST00000543627.1
|
DDB1
|
damage-specific DNA binding protein 1, 127kDa |
| chr3_-_111852061 | 0.13 |
ENST00000488580.1
ENST00000460387.2 ENST00000484193.1 ENST00000487901.1 |
GCSAM
|
germinal center-associated, signaling and motility |
| chr1_+_155583012 | 0.13 |
ENST00000462250.2
|
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr20_+_30327063 | 0.13 |
ENST00000300403.6
ENST00000340513.4 |
TPX2
|
TPX2, microtubule-associated |
| chr10_-_70092671 | 0.13 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr7_-_102789629 | 0.13 |
ENST00000417955.1
ENST00000341533.4 ENST00000425379.1 |
NAPEPLD
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr2_-_61244550 | 0.13 |
ENST00000421319.1
|
PUS10
|
pseudouridylate synthase 10 |
| chr11_-_33774944 | 0.12 |
ENST00000532057.1
ENST00000531080.1 |
FBXO3
|
F-box protein 3 |
| chr8_-_63998590 | 0.12 |
ENST00000260116.4
|
TTPA
|
tocopherol (alpha) transfer protein |
| chr12_+_12510045 | 0.12 |
ENST00000314565.4
|
LOH12CR1
|
loss of heterozygosity, 12, chromosomal region 1 |
| chr19_-_43422019 | 0.12 |
ENST00000402603.4
ENST00000594375.1 |
PSG6
|
pregnancy specific beta-1-glycoprotein 6 |
| chr8_+_7705398 | 0.12 |
ENST00000400125.2
ENST00000434307.2 ENST00000326558.5 ENST00000351436.4 ENST00000528033.1 |
SPAG11A
|
sperm associated antigen 11A |
| chr1_-_169337176 | 0.12 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr18_+_3247779 | 0.12 |
ENST00000578611.1
ENST00000583449.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr12_+_69753448 | 0.12 |
ENST00000247843.2
ENST00000548020.1 ENST00000549685.1 ENST00000552955.1 |
YEATS4
|
YEATS domain containing 4 |
| chr8_-_7320974 | 0.12 |
ENST00000528943.1
ENST00000359758.5 ENST00000361111.2 ENST00000398462.2 ENST00000297498.2 ENST00000317900.5 |
SPAG11B
|
sperm associated antigen 11B |
| chr4_-_75695366 | 0.12 |
ENST00000512743.1
|
BTC
|
betacellulin |
| chr1_+_42928945 | 0.12 |
ENST00000428554.2
|
CCDC30
|
coiled-coil domain containing 30 |
| chr14_+_39734482 | 0.12 |
ENST00000554392.1
ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5
|
CTAGE family, member 5 |
| chr7_-_2349579 | 0.12 |
ENST00000435060.1
|
SNX8
|
sorting nexin 8 |
| chr13_-_46716969 | 0.12 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr5_+_154238149 | 0.12 |
ENST00000519430.1
ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr3_-_187455680 | 0.12 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr12_+_62860581 | 0.12 |
ENST00000393632.2
ENST00000393630.3 ENST00000280379.6 ENST00000546600.1 ENST00000552738.1 ENST00000393629.2 ENST00000552115.1 |
MON2
|
MON2 homolog (S. cerevisiae) |
| chr12_-_88535747 | 0.11 |
ENST00000309041.7
|
CEP290
|
centrosomal protein 290kDa |
| chr20_+_814377 | 0.11 |
ENST00000304189.2
ENST00000381939.1 |
FAM110A
|
family with sequence similarity 110, member A |
| chr1_-_10532531 | 0.11 |
ENST00000377036.2
ENST00000377038.3 |
DFFA
|
DNA fragmentation factor, 45kDa, alpha polypeptide |
| chr17_+_30469579 | 0.11 |
ENST00000354266.3
ENST00000581094.1 ENST00000394692.2 |
RHOT1
|
ras homolog family member T1 |
| chr4_+_52917451 | 0.11 |
ENST00000295213.4
ENST00000419395.2 |
SPATA18
|
spermatogenesis associated 18 |
| chrX_-_48693955 | 0.11 |
ENST00000218230.5
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr1_-_94344754 | 0.11 |
ENST00000436063.2
|
DNTTIP2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr13_+_25875662 | 0.11 |
ENST00000381736.3
ENST00000463407.1 ENST00000381718.3 |
NUPL1
|
nucleoporin like 1 |
| chr1_-_85358850 | 0.11 |
ENST00000370611.3
|
LPAR3
|
lysophosphatidic acid receptor 3 |
| chr4_-_106629796 | 0.11 |
ENST00000416543.1
ENST00000515819.1 ENST00000420368.2 ENST00000503746.1 ENST00000340139.5 ENST00000433009.1 |
INTS12
|
integrator complex subunit 12 |
| chr11_+_124824000 | 0.11 |
ENST00000529051.1
ENST00000344762.5 |
CCDC15
|
coiled-coil domain containing 15 |
| chr18_+_21033239 | 0.11 |
ENST00000581585.1
ENST00000577501.1 |
RIOK3
|
RIO kinase 3 |
| chr4_+_113558272 | 0.11 |
ENST00000509061.1
ENST00000508577.1 ENST00000513553.1 |
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr10_+_105881906 | 0.10 |
ENST00000369727.3
ENST00000336358.5 |
SFR1
|
SWI5-dependent recombination repair 1 |
| chr15_+_41186637 | 0.10 |
ENST00000558474.1
|
VPS18
|
vacuolar protein sorting 18 homolog (S. cerevisiae) |
| chr3_-_57678772 | 0.10 |
ENST00000311128.5
|
DENND6A
|
DENN/MADD domain containing 6A |
| chr19_+_44716768 | 0.10 |
ENST00000586048.1
|
ZNF227
|
zinc finger protein 227 |
| chr5_+_134240588 | 0.10 |
ENST00000254908.6
|
PCBD2
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 |
| chr9_-_6015607 | 0.10 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr7_+_138818490 | 0.10 |
ENST00000430935.1
ENST00000495038.1 ENST00000474035.2 ENST00000478836.2 ENST00000464848.1 ENST00000343187.4 |
TTC26
|
tetratricopeptide repeat domain 26 |
| chr12_-_46121554 | 0.10 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr18_-_59274139 | 0.10 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chrX_-_153979315 | 0.10 |
ENST00000369575.3
ENST00000369568.4 ENST00000424127.2 |
GAB3
|
GRB2-associated binding protein 3 |
| chr4_+_187148556 | 0.10 |
ENST00000264690.6
ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1 |
| chr6_+_73331520 | 0.10 |
ENST00000342056.2
ENST00000355194.4 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr3_-_57233966 | 0.10 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr12_-_48597170 | 0.10 |
ENST00000310248.2
|
OR10AD1
|
olfactory receptor, family 10, subfamily AD, member 1 |
| chr1_+_28052456 | 0.10 |
ENST00000373954.6
ENST00000419687.2 |
FAM76A
|
family with sequence similarity 76, member A |
| chr6_+_53976235 | 0.10 |
ENST00000502396.1
ENST00000358276.5 |
MLIP
|
muscular LMNA-interacting protein |
| chr20_+_9049303 | 0.10 |
ENST00000407043.2
ENST00000441846.1 |
PLCB4
|
phospholipase C, beta 4 |
| chr2_+_46926048 | 0.10 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr13_-_41706864 | 0.10 |
ENST00000379485.1
ENST00000499385.2 |
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr12_+_123459127 | 0.10 |
ENST00000397389.2
ENST00000538755.1 ENST00000536150.1 ENST00000545056.1 ENST00000545612.1 ENST00000538628.1 ENST00000545317.1 |
OGFOD2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr4_-_113558014 | 0.10 |
ENST00000503172.1
ENST00000505019.1 ENST00000309071.5 |
C4orf21
|
chromosome 4 open reading frame 21 |
| chr15_-_85197501 | 0.10 |
ENST00000434634.2
|
WDR73
|
WD repeat domain 73 |
| chr1_-_111991850 | 0.10 |
ENST00000411751.2
|
WDR77
|
WD repeat domain 77 |
| chr11_+_112047087 | 0.10 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr2_+_73114489 | 0.10 |
ENST00000234454.5
|
SPR
|
sepiapterin reductase (7,8-dihydrobiopterin:NADP+ oxidoreductase) |
| chr2_+_109335929 | 0.10 |
ENST00000283195.6
|
RANBP2
|
RAN binding protein 2 |
| chr7_-_95025661 | 0.10 |
ENST00000542556.1
ENST00000265627.5 ENST00000427422.1 ENST00000451904.1 |
PON1
PON3
|
paraoxonase 1 paraoxonase 3 |
| chr3_-_146262428 | 0.10 |
ENST00000486631.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr15_+_74919779 | 0.09 |
ENST00000566926.1
|
CLK3
|
CDC-like kinase 3 |
| chr1_+_209859510 | 0.09 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr9_+_131901661 | 0.09 |
ENST00000423100.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr2_+_196522032 | 0.09 |
ENST00000418005.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr9_-_130889990 | 0.09 |
ENST00000449878.1
|
PTGES2
|
prostaglandin E synthase 2 |
| chr14_-_31926623 | 0.09 |
ENST00000356180.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr19_+_58193337 | 0.09 |
ENST00000601064.1
ENST00000282296.5 ENST00000356715.4 |
ZNF551
|
zinc finger protein 551 |
| chr9_+_96793076 | 0.09 |
ENST00000375360.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr17_-_5487768 | 0.09 |
ENST00000269280.4
ENST00000345221.3 ENST00000262467.5 |
NLRP1
|
NLR family, pyrin domain containing 1 |
| chr5_-_156569850 | 0.09 |
ENST00000524219.1
|
HAVCR2
|
hepatitis A virus cellular receptor 2 |
| chr12_+_121647868 | 0.09 |
ENST00000359949.7
ENST00000541532.1 ENST00000543171.1 ENST00000538701.1 |
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr4_-_169931393 | 0.09 |
ENST00000504480.1
ENST00000306193.3 |
CBR4
|
carbonyl reductase 4 |
| chr1_-_118727781 | 0.09 |
ENST00000336338.5
|
SPAG17
|
sperm associated antigen 17 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HSFY2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.3 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.2 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.1 | 0.2 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.1 | 0.2 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.2 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.2 | GO:2000685 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.0 | 0.1 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.0 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.3 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.0 | 0.2 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.2 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.3 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.2 | GO:1903998 | response to isolation stress(GO:0035900) regulation of eating behavior(GO:1903998) negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.2 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.1 | GO:0002519 | natural killer cell tolerance induction(GO:0002519) regulation of tolerance induction dependent upon immune response(GO:0002652) negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.0 | 0.1 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.0 | 0.2 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.1 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:1904468 | negative regulation of tumor necrosis factor secretion(GO:1904468) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0090210 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.0 | 0.4 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.0 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.2 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.1 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.1 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.0 | 0.1 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 0.2 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.0 | GO:0050904 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.0 | GO:0060057 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.2 | GO:0015827 | tryptophan transport(GO:0015827) |
| 0.0 | 0.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.1 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.0 | 0.0 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.4 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.0 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.0 | 0.1 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.3 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 0.2 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.0 | 0.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0070685 | macropinocytic cup(GO:0070685) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.0 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 0.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.3 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.1 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0008124 | phenylalanine 4-monooxygenase activity(GO:0004505) 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.3 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.1 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.0 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.3 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.2 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |